1000/1000
Hot
Most Recent

| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Herman Spaink | + 2808 word(s) | 2808 | 2021-02-25 04:10:42 | | | |
| 2 | Peter Tang | Meta information modification | 2808 | 2021-02-27 12:51:30 | | |
The genus Mycobacterium comprises a multitude of species known to cause serious disease in humans, including Mycobacterium tuberculosis and M. leprae, the responsible agents for tuberculosis and leprosy, respectively. In addition, there is a worldwide spike in the number of infections caused by a mixed group of species such as the M. avium, M. abscessus and M. ulcerans complexes, collectively called nontuberculous mycobacteria (NTMs). The situation is forecasted to worsen because, like tuberculosis, NTMs either naturally possess or are developing high resistance against conventional antibiotics.
Mycobacteria are a large group of non-motile, rod-shaped bacteria that tend to grow mold-like pellicles on liquid culture media. Out of the 150 species known to this genus, nearly 25 are known to cause disease in humans. The most well-known mycobacteria species are the M. tuberculosis and M. leprae complexes, with an estimated prevalence rate of 130 (the year 2020) and 2 (the year 2018) cases per 100,000 population, respectively [1][2], while all others are collectively called nontuberculous mycobacteria (NTMs) [3]. Despite that NTMs are less widespread pathogens for humans than M. tuberculosis, they have proven to be an emerging threat to the immunocompromised population [4], with an estimated 4.1–14.1 cases per 100,000 population worldwide (2013) [5]. NTMs are ubiquitous and can survive in a wide range of environmental conditions, and their infections are difficult to diagnose [6]. The most common NTM-related pathologies are pulmonary infections (pulmonary nontuberculous mycobacterial disease) caused by strains from the M. avium complex and M. abscessus [6][7], but NTMs can also cause skin and soft tissue infections (e.g., M. marinum infection and Buruli ulcer caused by M. ulcerans), lymphadenitis in immunocompromised children, and even invasive disseminated disease eventually leading to death.
According to Runyon, NTMs can be classified based on the growth rate and pigment formation (Table 1) [8]. Types I, II, and III strains are classified as slow-growers because they take seven or more days of growth for forming visible colonies on a subculture plate [9]. They are differentiated on their ability to produce pigments only on exposure to light (type I or photochromogens) or also in the dark (type II or scotochromogens), or not being strongly pigmented (type III or non-photochromogens) [10]. Type IV strains are regarded as rapid-growers as they take less than seven days to form visible colonies on a subculture plate [10]. Generally, slow-growing mycobacteria are much more prevalent than fast-growing ones [11] and present higher ratios of drug resistance (with the fast-growing M. abscessus being a notable exception) [12]. It has been suggested that all mycobacteria evolved from a common ancestral rapid growing mycobacterial strain [13][14][15].
Table 1. Summary of the nontuberculous mycobacteria (NTMs) mentioned in this review, their classification according to Runyon, and their reported pathogenesis in humans.
|
Runyon Classification |
NTM Species |
Pathogenesis in Humans |
|---|---|---|
|
Photochromogens Runyon type I |
M. kansasii [16], M. simiae [17] |
Pulmonary infections Skin infections Disseminated infections |
|
M. marinum [18] |
Skin and soft tissue infections Disseminated infections |
|
|
Scotochromogens Runyon type II |
M. gordonae [19] |
Pulmonary infections Skin infections Disseminated infections |
|
M. scrofulaceum [20] |
Cervical lymphadenitis among children Pulmonary infections Disseminated infections |
|
|
Non-photochromogens Runyon type III |
M. avium complex (M. avium and M. intracellulare) [21] |
Pulmonary MAC infections Disseminated infections (mostly in AIDS patients) MAC associated lymphadenitis (in young kids and people with normal immune systems) |
|
M. malmoense [22] |
Pulmonary infections Disseminated infections |
|
|
M. ulcerans [18] |
Skin diseases (Buruli ulcers) |
|
|
Rapid growing Runyon type IV |
M. abscessus [23] |
Pulmonary infections Skin and Soft tissue disease Central nervous system infections Disseminated infections |
|
M. chelonae [24] |
Skin and soft tissue infections Pulmonary infections Disseminated infections |
|
|
M. smegmatis |
Widely regarded as nonpathogenic |
Recently, there has been a considerable increase in the number of reported NTM related diseases, including respiratory infections caused by various strains from the M. avium complex, M. kansasii and M. abscessus [25]. This is partly because of the awareness of the symptoms caused by these infections and improvements in detection techniques, but also because of an increase in the number of susceptible individuals and that NTM can form biofilms in common household and hospital sources of infection (such as showerheads, faucets, water distribution systems, plumbing systems, etc.) [26][27]. The situation is worrying because, just like tuberculosis, these bacteria have developed high resistance against conventional antibiotics [28]. However, these pathogens are still considered opportunistic since they require a combination of constant exposure as well as host susceptibility to infection, and these infections have mainly remained limited to patients with pre-existing lung diseases [25][29].
The major NTM that is infecting such individuals suffering from chronic diseases like cystic fibrosis is M. abscessus, which is a rapidly growing, intrinsically multidrug-resistant species [30]. These infections are often impossible to treat despite prolonged antibiotic therapy, and the therapy may even be contraindicated with lung transplantation, leaving no effective options for treatment [31]. While NTM infections were earlier thought to be independently acquired by susceptible individuals, the recent consensus is that such infections are frequently transmitted indirectly from an infected to a healthy individual, for instance, via contaminated hospital equipment [32]. Some opportunistic infectious NTM species tend to cluster in specific geographical distributions, and there may be a genetic basis for the susceptibility to their infection in particular patients [11][33][34]. Finally, relapse and reinfection is a major problem with some NTM infections, like the ones caused by M. avium complex [35], although it is less so for other species like M. kansasii [36].
Currently, the treatment for almost all NTM infections is based on macrolide-based antibiotics, such as clarithromycin or azithromycin. For NTM infections caused by the slow-growing group, the regime also includes ethambutol and rifampicin [37], while for fast-growers, it includes an aminoglycoside and either cefoxitin, imipenem or tigecycline [38]. These treatments are largely empirical, derived from years of clinical practice, can last for as long as 18 months, are costly, and are often associated with drug-related toxicities and side-effects [39]. Cure rates range from 80–90% with M. malmoense infections to just 30–50% with M. abscessus infections [40]. Thus, the discovery of new and more efficient therapies against NTMs is an important topic of research. However, a major bottleneck is the low susceptibility of mycobacteria to most antibiotics, including the ones used against tuberculosis [41]. A better understanding of the underlying mechanisms behind this drug resistance by improving the available models to study their infection could significantly help in accelerating the drug discovery process.
Drug Resistance can be either intrinsic (natural) or acquired [42]. Intrinsic resistance describes a situation where an organism possesses a set of special features that allows it to tolerate a particular drug or survive in an otherwise hostile chemical environment [42]. Mechanisms by which NTMs are intrinsically resistant to antibiotics include their thick, impermeable cell walls or their presence in biofilms and granulomas, which effectively decrease drug uptake, as well as the expression of proteins that specifically target clinically used antibacterial compounds.
On the other hand, acquired resistance refers to the case where a resistant strain emerges from a population that was previously drug-sensitive [42]. These events are usually related to the prolonged antibiotic treatments required to cure NTM infections. The acquired resistance is particularly severe for NTMs that only have a single copy of genes encoding common target proteins such as ribosomes, thus increasing the risk of acquiring protective mutations with single-drug treatments [4][43]. Here, we will focus on the mechanisms of mycobacterial physiology that make them naturally resistant to antimicrobial treatments since Nasiri et al. recently reviewed the mutations that may cause resistance to certain antibiotics in NTM [44].
Conceptually, resistance to antimicrobial drugs can be a result of one or more of the following mechanisms: decreased drug uptake, increased drug efflux, increased drug metabolism, or reduced drug sequestration (Figure 1) [45].
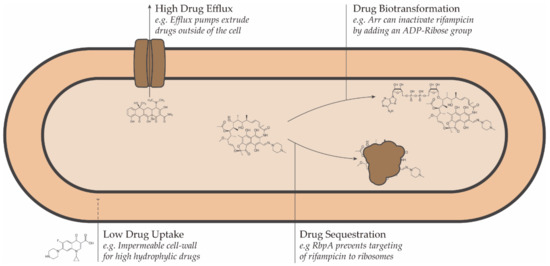
Figure 1. Schematic representation of the intrinsic drug resistance mechanisms in bacteria.
There is a wide range of techniques that can be employed in the development of new potential antimicrobial therapies against NTMs. These techniques can be classified depending on the tools employ between in silico, in vitro, or in vivo. In general terms, in silico techniques are useful to generate new leads and narrow the search of potential candidates based on prior information at the start of a study or to optimize compounds based on specific targets via virtual simulations. These leads can then be tested for efficacy using standardized in vitro analysis, which allows the determination of their potential antimycobacterial activity. Finally, in vivo animal models can be used to recreate infection environments and are therefore interesting for preclinical evaluation of potential compounds. We summarized the main attributes for each category in Table 2. A recent review by Rampacci et al. explains in detail the different techniques, assays, and preclinical models against NTMs that have been developed, with an emphasis on the newer models [46]. We direct the reader to that review for an in-depth description of these methodologies and their read-outs. Here, we will give a brief outline of the most common techniques implemented in the lab and how they complement each other to create an integrated pipeline for drug discovery.
Table 2. Summary of the current methods available for the discovery of new antimicrobial therapies in NTMs.
|
In Silico |
In Vitro |
In Vivo |
|
|---|---|---|---|
|
Methods employed |
Structure–activity relations, Molecular simulations, Comparative genomics |
Antimicrobial effect tests on cultured cells |
Tests on live infected animals |
|
Main insights |
Molecular basis for drug action |
Molecular and cellular effect of drug action |
Whole-organism level of drug action |
|
Advantages |
High throughput, low-cost, no need for actual chemical synthesis of compounds or bacterial growth |
Relatively simple systems and lower cost and time involvement, easy to handle, scalable |
Closer to the actual physiological environment |
|
Limitations |
Requires prior information and complicated models to simulate molecular events such as docking and drug-target interactions |
Needs a high level of standardization and careful experimentation for reproducibility, may not reproduce clinical situations |
Requires careful model selection, large organism response is less predictable, ethical considerations, high economical costs |
|
Best-fit stage in drug discovery |
Primary (for narrowing the search of potential candidates) or secondary (for optimizing compounds to species-specific targets) |
Secondary (for screening initial targets and efficacy determination) |
Tertiary (for preclinical evaluation) |
When designing new possible therapies against NTMs, it is important to consider the mechanisms that confer drug resistance that we reviewed above. For example, the exceptionally high hydrophobicity of mycobacterial cell walls has an important bearing on drug design: the more lipophilic molecules generally show higher permeability and hence are more active. This means that a possible route to developing anti-NTM antibiotics is to synthesize hydrophobic derivatives of existing antibiotics [47]. For instance, ciprofloxacin, when modified by the addition of hydrophobic alkyl substituents, showed higher activity against M. avium [48]. Similarly, for M. leprae, the efficacy of fluoroquinolones improved by incorporating the hydrophobic cyclopropyl groups [49].
Compounds that have been identified to be active against other diseases may directly be screened by in vitro bacterial assays, and MIC values may be determined to check the efficiency of the drug [50]. However, for M. abscessus, the hit rates among drug libraries that are active against neglected diseases like ascariasis, Buruli ulcer, Chagas disease, and malaria is just 1% [51], highlighting the great difficulty in finding new drugs for NTMs. A way forward could be to screen the compound libraries active against tuberculosis for their effect against NTM because of the structural similarity and homology of their drug targets [52]. In a recently conducted study, 129 compounds known to be active against M. tuberculosis were tested against M. abscessus and M. avium, and their rates were higher than for drugs that are not active against tuberculosis [53]. Rifabutin, an antibiotic used for the treatment of tuberculosis, has recently been found to be active against M. abscessus [50]. Notwithstanding these positive outcomes, most existing drugs specific to tuberculosis are usually ineffective against NTM [53][54].
In the development of a novel treatment, it should be considered that most of the successful anti-NTM drug therapies involve synergistic effects of two drugs: one antibiotic to disrupt the permeability of the outer membrane in order to ensure entry of the drug into the cell, and the another disrupting at least one vital cellular processes (such as DNA, RNA, or protein and outer membrane synthesis) for inhibiting cell growth [55]. For example, the performance of hydrophobic drugs that have intracellular targets can be improved by using them in conjunction with compounds that specifically target cell wall homeostasis. Such synergistic effects have been observed between ethambutol plus rifampicin in M. avium [56] or vancomycin plus clarithromycin in M. abscessus [57]. Similar effects can be seen using adjuvants that inhibit specific efflux pumps [58][59] or that increase the expression of the enzymes required for the biotransformation of a prodrug, thus boosting antibiotic effectivity [60].
Moreover, the synergistic effects cannot only improve drug efficacy but also reduce the chances that treatments lead to drug resistance [61][62]. For instance, the combination of β-lactam antibiotics and β-lactamase inhibitors has shown to be a promising strategy against M. avium infections [63]. Further, synergies can also be achieved by using a combination of two or more drugs that have the same cellular target. This approach has been validated for M. abscessus complex, wherein a dual β-lactam drug regimen proved to be much more effective than a single-drug regimen, with or without β-lactamase inhibitors [64][65]. In such regimens, each of the β-lactams preferentially targets a different enzyme that is involved in cell wall synthesis, thereby ensuring that “overlapping” effects are minimized, and combinedly, all biochemical pathways are exhaustively targeted [65]. Typically, mutations that cause the development of resistance mechanisms can have subsequent “spill-over” effects: the same mechanism may confer resistance to the entire class of drugs that target the same biochemical pathway (cross-resistance), or it can lead to increased vulnerability to other drugs that target a different pathway (collateral sensitivity) [66]. Exploiting collateral sensitivity by either combinatorial or cyclical treatment regimens involving multiple drugs can be an effective strategy, as recently demonstrated with drugs directed against M. marinum [67].
A different approach towards finding more effective treatments against NTM infections is host-directed therapies (HDT), in which specific immune pathways of the host are modulated in such a way that it leads to a better clinical outcome. That is, HDTs aims to empower the host to clear the infection instead of directly targeting bacteria. The ways in which HDTs can help against NTMs are strengthening innate immunity against mycobacterial infections, preventing the growth of the bacilli by inhibiting the essential host-related growth-factors, restoring the immune response suppressed due to the infection, or reducing tissue damage due to hyperinflammation [68][69].
HDTs offer several unique advantages over conventional therapies. First, chances of drug-related resistance are considerably reduced because it is difficult for the bacteria to develop completely new mechanisms of interacting with the host quickly and while being under the same hostile immune selection [70]. They also offer the possibility of making conventional drugs more effective against already resistant strains by neutralizing pathogen defenses [71]. The synergistic role of HDT adjuvants with anti-mycobacterials was demonstrated in a study in which picolinic acid (PA) was shown to potentiate fluoroquinolones against bacteria from the M. avium complex [72]. Fluoroquinolones are otherwise only very weakly effective against M. avium infections. This was attributed to two factors- upregulation of the immune system by PA and chelation of Fe ions by PA, which deprive the bacteria of the essential ions needed for growth [73]. In a previous study, PA was shown to inhibit M. avium growth inside mouse macrophages by inducing apoptosis- causing morphological changes [74].
In addition, some compounds show both host-directed and bacterial-directed actions. Clofazimine, a commonly used drug in M. leprae infections, is a good example of a drug that simultaneously affects the host as well as the bacteria. Upon infecting the body, M. leprae creates a safe microenvironment for itself inside the macrophages of the host by increasing the accumulation and retarding the breakdown of macrophage lipids [75]. It was shown that clofazimine not only helps reverse these two processes but also activates immune reactions in M. leprae infected host cells. Hence, effectively, it not just prevents the growth of the bacteria but also actively helps to eliminate it [76]. Another example of an antibiotic with a strong effect on the host inflammatory system is minocycline that modulates the endocannabinoid signaling pathway and, in this way, might have HDT potential [77].
Finally, considering the mechanisms of defense of the host system can lead to more effective ways of delivering drugs to the target, adding value to existing treatments. An example would be precision-targeting the drug by loading them into the host cells that act as carriers. This approach was demonstrated by loading dendritic cells to deliver amikacin inside alveolar granulomas and thus enhancing the killing of residing mycobacteria [78].
However, a major challenge in the development of effective HDTs is that different patients may not have the same immune status, which may depend on factors like the stage of the disease, health of the individual, pre-existing conditions, and genetic makeup [71]. This can be a hindrance towards a universal HDT and may require an approach for personalized medicine, much like cancer immunotherapy [79].