
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Marcio Dorn | -- | 2836 | 2023-02-16 15:22:59 | | | |
| 2 | Lindsay Dong | Meta information modification | 2836 | 2023-02-17 06:18:53 | | |
Video Upload Options
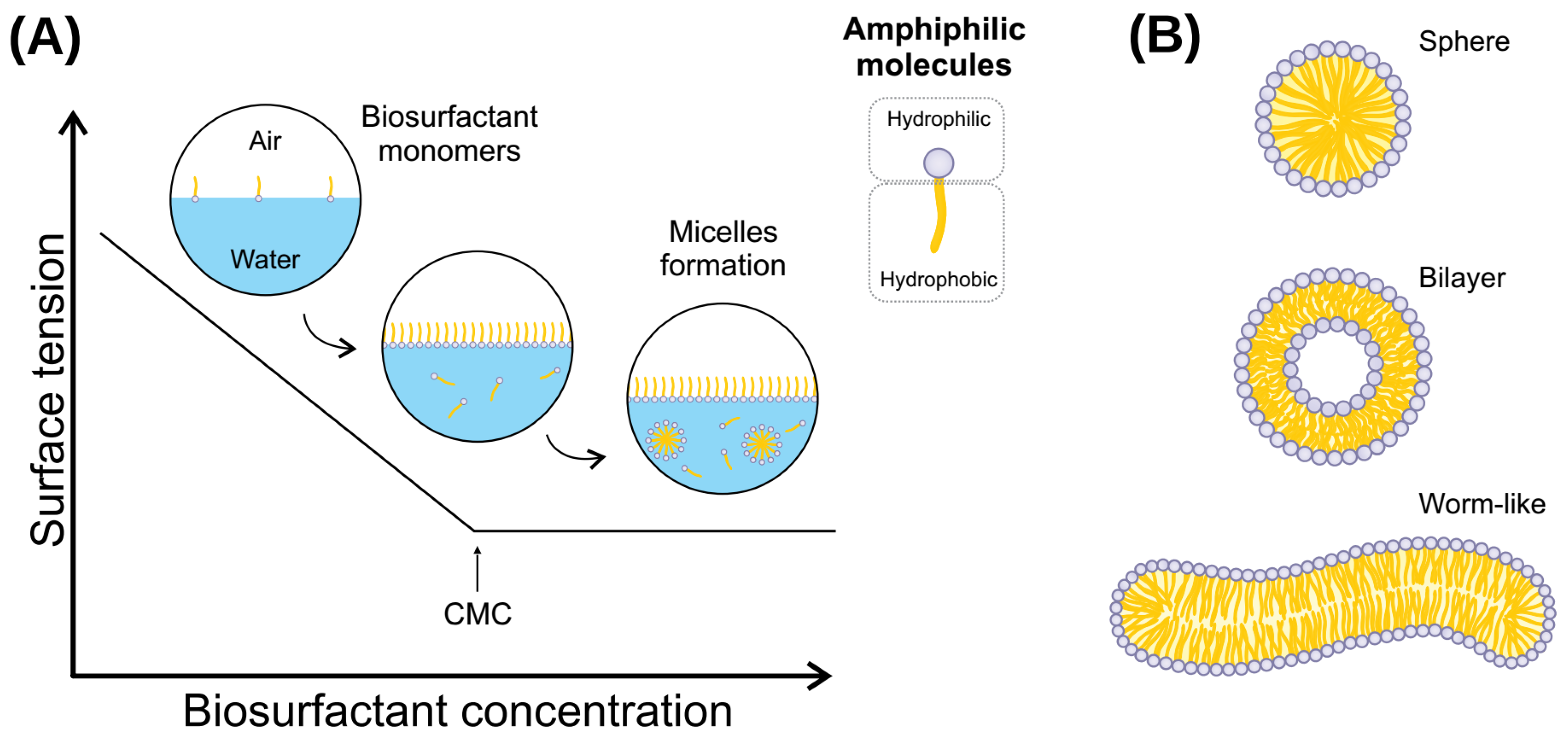
Biosurfactants are amphipathic molecules capable of lowering interfacial and superficial tensions. Produced by living organisms, these compounds act the same as chemical surfactants but with a series of improvements, the most notable being biodegradability. Biosurfactants have a wide diversity of categories. Within these, lipopeptides are some of the more abundant and widely known.
1. Introduction
2. Biosurfactant-Producing Microorganisms
2.1. Lipopeptides
2.2. Protein-Containing Biosurfactants
3. Lipopeptides Synthesis and Regulation
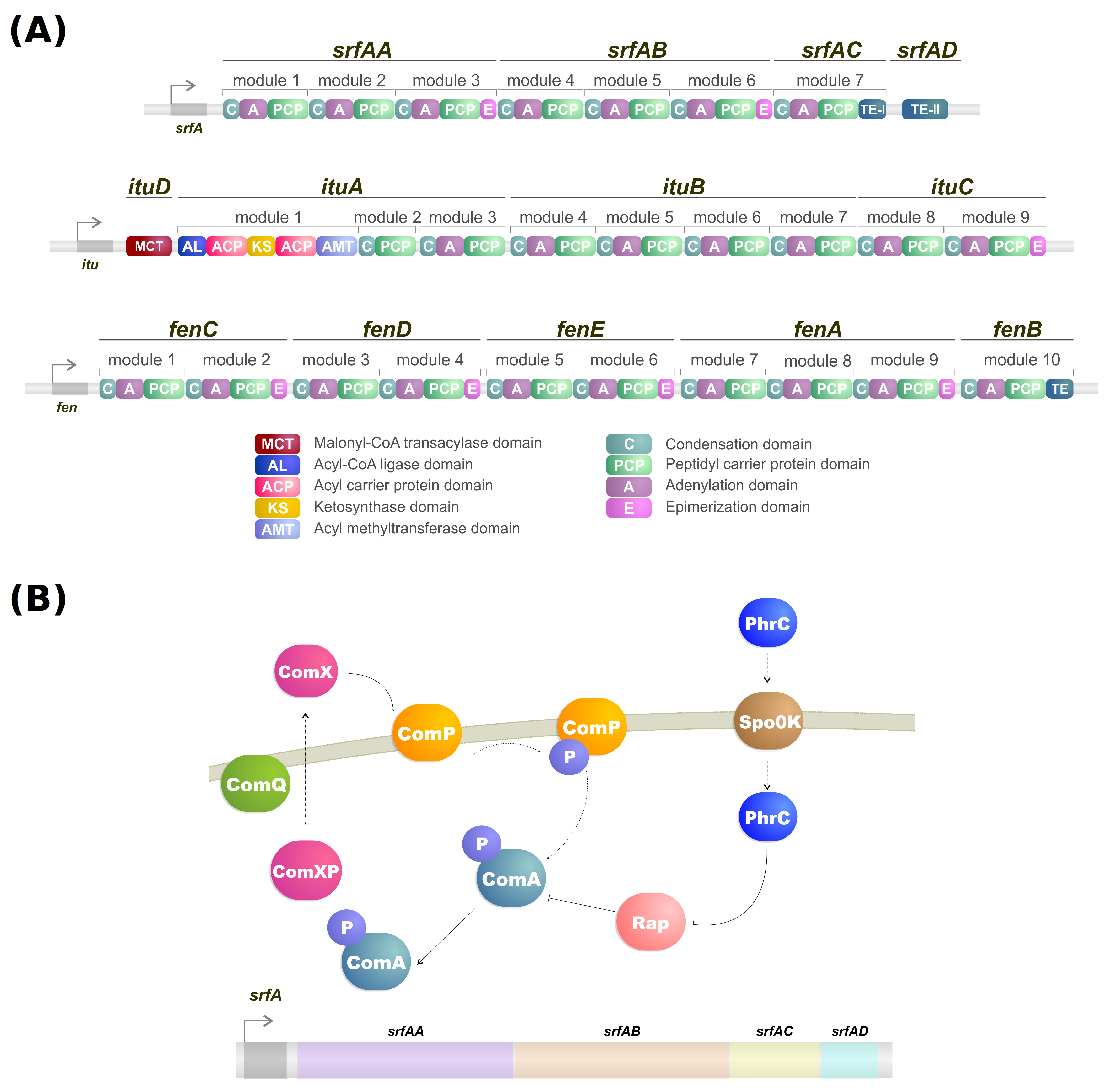
NRPSs are multidomain mega-enzymes that synthesize NRPs without using the ribosomal machinery [24]. They have a modular structure in which each part incorporates an amino acid to the peptide moiety of the lipopeptide (Figure 1B). Another characteristic is that the NRPS obeys the colinearity rule; that is, the modules are colinear with the amino acid sequence of the peptide [16]. There are two types of modules: initiation and elongation modules. Each of these modules also contains domains that perform specific tasks [16]. While typically, initiation modules have domains responsible for the amino acid election, activation, and thioesterification of the activated amino acid, the first module also contains a condensation domain in lipopeptide biosynthesis. This domain is responsible for catalyzing the N-acylation of the first amino acid of the lipopeptide, thus linking the lipid moiety to the oligopeptide [16][25]. Elongation modules contain the same domains, but this time the condensation domain is responsible for catalyzing the peptide bond between two amino acids. The condensation domain from the elongation module will generate a lipopeptide that, by the end of the assembly line, will be cleaved by a thioesterase [16].
It has been shown via analyses of the metabolic profiles of Pseudomonas and Bacillus species that a single strain may be able to produce different forms of the same biosurfactant [16]. Examples include B. subtilis strain OKB 105, which can produce 12 different surfactin analogs [26] and P. fluorescens strain SS101 that can produce up to eight analogs of the lipopeptide massetolide A [27]. These analogs are thought to be the product of flexibility in the amino acid selection and activation by the adenylation domain of the initiation module in NRPSs. Said flexibility is common in nonribosomal peptide synthesis, which may have biological purposes for the producers [16].
Iturin production has a series of regulation factors. For example, the methylation of tyrosine residues in iturin can decerease yield and antibacterial activity [28][29]. Furthermore, iturin can also be regulated by controlling the expression of sigma factor A and the transcription factor ComA. Overexpressing the genes sigA and comA increased iturin yield [30].
As mentioned, the biosynthesis of surfactin is coordinated by the srfA operon, which contains four open reading frames (srfAA, srfAB, srfAC, and srfAD) as seen in Figure 1A [31]. These open reading frames are responsible for the peptide chain extension, a key step in the surfactin synthesis regulation [29]. Surfactin, much like iturin, is also strongly regulated by the transcription factor ComA, which binds to srfA, and thus controls its transcription and regulates srfA expression (Figure 1B) [29]. ComA phosphorylation is the key to activating the srfA operon transcription by two different pathways. The first involves the ComX peptide modified by ComQ that stimulates ComP autophosphorylation, triggering ComA phosphorylation (Figure 1A) [32]. Activated ComA translocates to the nucleus and promotes srfA transcription. PhrC importation is mediated by Spo0K, which interacts with Rap protein and inhibits its phosphatase activity, thus, preventing ComA dephosphorylation and facilitating ComA-induced srfA transcription (Figure 1A) [32][33].
4. Biosurfactant Toxicity
5. Emerging Strategies for Biosurfactant Production
6. Physical and Chemical Properties of Biosurfactants
7. Omics Technology and Bioinformatic Analysis as a Tool for Biosurfactant Identification
7.1. Network Analysis
7.2. Machine Learning and Data Integration
7.3. Molecular Dynamics (MD) Simulations
References
- Jahan, R.; Bodratti, A.M.; Tsianou, M.; Alexandridis, P. Biosurfactants, natural alternatives to synthetic surfactants: Physicochemical properties and applications. Adv. Colloid Interface Sci. 2020, 275, 102061.
- Hamme, J.D.V.; Singh, A.; Ward, O.P. Physiological aspects: Part 1 in a series of papers devoted to surfactants in microbiology and biotechnology. Biotechnol. Adv. 2006, 24, 604–620.
- Singh, P.; Patil, Y.; Rale, V. Biosurfactant production: Emerging trends and promising strategies. J. Appl. Microbiol. 2019, 126, 2–13.
- Desai, J.D.; Banat, I.M. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 1997, 61, 47–64.
- Machlin, E.S. CHAPTER XIV—Thermodynamics of Micelles. In An Introduction to Aspects of Thermodynamics and Kinetics Relevant to Materials Science, 3rd ed.; Machlin, E.S., Ed.; Elsevier Science Ltd.: Oxford, UK, 2007; pp. 425–454.
- Cameotra, S.S.; Makkar, R.S.; Kaur, J.; Mehta, S.K. Synthesis of Biosurfactants and Their Advantages to Microorganisms and Mankind. In Biosurfactants; Sen, R., Ed.; Springer: New York, NY, USA, 2010; pp. 261–280.
- Kiran, G.S.; Sabarathnam, B.; Selvin, J. Biofilm disruption potential of a glycolipid biosurfactant from marine Brevibacterium casei. FEMS Immunol. Med. Microbiol. 2010, 59, 432–438.
- Franzetti, A.; Gandolfi, I.; Fracchia, L.; Van Hamme, J.; Gkorezis, P.; Marchant, R.; Banat, I.M. Biosurfactant use in heavy metal removal from industrial effluents and contaminated sites. Biosurfactants Prod. Util. Technol. Econ. 2014, 159, 361.
- Reis, R.; Pacheco, G.; Pereira, A.; Freire, D. Biosurfactants: Production and Applications. In Biodegradation; Chamy, R., Rosenkranz, F., Eds.; IntechOpen: Rijeka, Croatia, 2013; Chapter 2.
- Souza, K.S.T.; Gudiña, E.J.; Schwan, R.F.; Rodrigues, L.R.; Dias, D.R.; Teixeira, J.A. Improvement of biosurfactant production by Wickerhamomyces anomalus CCMA 0358 and its potential application in bioremediation. J. Hazard. Mater. 2018, 346, 152–158.
- Vijayakumar, S.; Saravanan, V. Biosurfactants-Types, Sources and Applications. Res. J. Microbiol. 2015, 10, 181–192.
- Roy, A. A Review on the Biosurfactants: Properties, Types and its Applications. J. Fundam. Renew. Energy Appl. 2017, 8, 1–14.
- Varjani, S.J.; Upasani, V.N. Critical review on biosurfactant analysis, purification and characterization using rhamnolipid as a model biosurfactant. Bioresour. Technol. 2017, 232, 389–397.
- Henkel, M.; Hausmann, R. Chapter 2—Diversity and Classification of Microbial Surfactants. In Biobased Surfactants, 3rd ed.; Hayes, D.G., Solaiman, D.K., Ashby, R.D., Eds.; AOCS Press: London, UK, 2019; pp. 41–63.
- Kashif, A.; Rehman, R.; Fuwad, A.; Shahid, M.K.; Dayarathne, H.; Jamal, A.; Aftab, M.N.; Mainali, B.; Choi, Y. Current advances in the classification, production, properties and applications of microbial biosurfactants—A critical review. Adv. Colloid Interface Sci. 2022, 2022, 102718.
- Raaijmakers, J.M.; De Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062.
- Marchant, R.; Funston, S.; Uzoigwe, C.; Rahman, P.; Banat, I.M. Production of Biosurfactants from Nonpathogenic Bacteria. In Biosurfactants: Production and Utilization-Processes, Technologies, and Economics; Kosaric, N., Sukan, F.V., Eds.; CRC Press: Boca Raton, FL, USA, 2014; pp. 73–81.
- Das, P.; Mukherjee, S.; Sen, R. Genetic regulations of the biosynthesis of microbial surfactants: An overview. Biotechnol. Genet. Eng. Rev. 2008, 25, 165–186.
- Carolin C, F.; Kumar, P.S.; Ngueagni, P.T. A review on new aspects of lipopeptide biosurfactant: Types, production, properties and its application in the bioremediation process. J. Hazard. Mater. 2021, 407, 124827.
- Long, X.; He, N.; He, Y.; Jiang, J.; Wu, T. Biosurfactant surfactin with pH-regulated emulsification activity for efficient oil separation when used as emulsifier. Bioresour. Technol. 2017, 241, 200–206.
- Schor, M.; Reid, J.L.; MacPhee, C.E.; Stanley-Wall, N.R. The Diverse Structures and Functions of Surfactant Proteins. Trends Biochem. Sci. 2016, 41, 610–620.
- Whitsett, J.A.; Weaver, T.E. Hydrophobic Surfactant Proteins in Lung Function and Disease. N. Engl. J. Med. 2002, 347, 2141–2148.
- Cirigliano, M.C.; Carman, G.M. Purification and characterization of liposan, a bioemulsifier from Candida lipolytica. Appl. Environ. Microbiol. 1985, 50, 846–850.
- Soltani, J. Chapter 22—Secondary Metabolite Diversity of the Genus Aspergillus: Recent Advances. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.K., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 275–292.
- Roongsawang, N.; Lim, S.P.; Washio, K.; Takano, K.; Kanaya, S.; Morikawa, M. Phylogenetic analysis of condensation domains in the nonribosomal peptide synthetases. FEMS Microbiol. Lett. 2005, 252, 143–151.
- Kowall, M.; Vater, J.; Kluge, B.; Stein, T.; Franke, P.; Ziessow, D. Separation and Characterization of Surfactin Isoforms Produced by B. subtilis OKB 105. J. Colloid Interface Sci. 1998, 204, 1–8.
- de Bruijn, I.; de Kock, M.J.D.; de Waard, P.; van Beek, T.A.; Raaijmakers, J.M. Massetolide A Biosynthesis in Pseudomonas fluorescens. J. Bacteriol. 2008, 190, 2777–2789.
- Moran, S.; Rai, D.K.; Clark, B.R.; Murphy, C.D. Precursor-directed biosynthesis of fluorinated iturin A in Bacillus spp. Org. Biomol. Chem. 2009, 7, 644–646.
- Yang, R.; Lei, S.; Xu, X.; Jin, H.; Sun, H.; Zhao, X.; Pang, B.; Shi, J. Key elements and regulation strategies of NRPSs for biosynthesis of lipopeptides by Bacillus. Appl. Microbiol. Biotechnol. 2020, 104, 8077–8087.
- Zhang, Z.; Ding, Z.; Zhong, J.; Zhou, J.; Shu, D.; Luo, D.; Yang, J.; Tan, H. Improvement of iturin A production in B. subtilis ZK 0 by overexpression of the comA and sigA genes. Lett. Appl. Microbiol. 2017, 64, 452–458.
- Cosmina, P.; Rodriguez, F.; de Ferra, F.; Grandi, G.; Perego, M.; Venema, G.; van Sinderen, D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in B. subtilis. Mol. Microbiol. 1993, 8, 821–831.
- Roongsawang, N.; Washio, K.; Morikawa, M. Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. Int. J. Mol. Sci. 2010, 12, 141–172.
- Hu, F.; Liu, Y.; Li, S. Rational strain improvement for surfactin production: Enhancing the yield and generating novel structures. Microb. Cell Factories 2019, 18, 42.
- Poremba, K.; Gunkel, W.; Lang, S.; Wagner, F. Marine Biosurfactants, III. Toxicity Testing with Marine Microorganisms and Comparison with Synthetic Surfactants. Z. Naturforschung C 1991, 46, 210–216.
- Ribeiro, B.G.; Guerra, J.M.C.; Sarubbo, L.A. Biosurfactants: Production and application prospects in the food industry. Biotechnol. Prog. 2020, 36, e3030.
- Edwards, K.R.; Lepo, J.E.; Lewis, M.A. Toxicity comparison of biosurfactants and synthetic surfactants used in oil spill remediation to two estuarine species. Mar. Pollut. Bull. 2003, 46, 1309–1316.
- Velioğlu, Z.; Ozturk Urek, R. Biosurfactant production by Pleurotus ostreatus in submerged and solid-state fermentation systems. Turk. J. Biol. 2015, 39, 160–166.
- Magro, A.E.A.; Silva, L.C.; Rasera, G.B.; de Castro, R.J.S. Solid-state fermentation as an efficient strategy for the biotransformation of lentils: Enhancing their antioxidant and antidiabetic potentials. Bioresour. Bioprocess. 2019, 6, 38.
- Cerda, A.; Artola, A.; Barrena, R.; Font, X.; Gea, T.; Sánchez, A. Innovative Production of Bioproducts from Organic Waste through Solid-State Fermentation. Front. Sustain. Food Syst. 2019, 3, 63.
- Das, K.; Mukherjee, A. Comparison of lipopeptide biosurfactants production by B. subtilis strains in submerged and solid state fermentation systems using a cheap carbon source: Some industrial applications of biosurfactants. Process. Biochem. 2007, 42, 1191–1199.
- Zhu, Z.; Zhang, G.; Luo, Y.; Ran, W.; Shen, Q. Production of lipopeptides by Bacillus amyloliquefaciens XZ-173 in solid state fermentation using soybean flour and rice straw as the substrate. Bioresour. Technol. 2012, 112, 254–260.
- Zouari, R.; Ellouze-Chaabouni, S.; Ghribi-Aydi, D. Optimization of B. subtilis SPB1 Biosurfactant Production under Solid-state Fermentation Using By-Products of a Traditional Olive Mill Factory. Achieve Life Sci. 2014, 8, 162–169.
- Jajor, P.; Piłakowska-Pietras, D.; Krasowska, A.; Łukaszewicz, M. Surfactin analogues produced by B. subtilis strains grown on rapeseed cake. J. Mol. Struct. 2016, 1126, 141–146.
- Kavuthodi, B.; Thomas, S.K.; Sebastian, D. Co-production of Pectinase and Biosurfactant by the Newly Isolated Strain B. subtilis BKDS1. Microbiol. Res. J. Int. 2015, 10, 1–12.
- Dexter, A.F.; Middelberg, A.P. Peptides as functional surfactants. Ind. Eng. Chem. Res. 2008, 47, 6391–6398.
- Marchant, R.; Banat, I.M. Microbial biosurfactants: Challenges and opportunities for future exploitation. Trends Biotechnol. 2012, 30, 558–565.
- Belhaj, A.F.; Elraies, K.A.; Mahmood, S.M.; Zulkifli, N.N.; Akbari, S.; Hussien, O.S. The effect of surfactant concentration, salinity, temperature, and pH on surfactant adsorption for chemical enhanced oil recovery: A review. J. Pet. Explor. Prod. Technol. 2020, 10, 125–137.
- De Almeida, D.G.; Soares Da Silva, R.d.C.F.; Luna, J.M.; Rufino, R.D.; Santos, V.A.; Banat, I.M.; Sarubbo, L.A. Biosurfactants: Promising molecules for petroleum biotechnology advances. Front. Microbiol. 2016, 7, 1718.
- Satpute, S.K.; Banpurkar, A.G.; Dhakephalkar, P.K.; Banat, I.M.; Chopade, B.A. Methods for investigating biosurfactants and bioemulsifiers: A review. Crit. Rev. Biotechnol. 2010, 30, 127–144.
- Bouchemal, K.; Agnely, F.; Koffi, A.; Djabourov, M.; Ponchel, G. What can isothermal titration microcalorimetry experiments tell us about the self-organization of surfactants into micelles? J. Mol. Recognit. 2010, 23, 335–342.
- Wu, S.; Liang, F.; Hu, D.; Li, H.; Yang, W.; Zhu, Q. Determining the critical micelle concentration of surfactants by a simple and fast titration method. Anal. Chem. 2019, 92, 4259–4265.
- Scholz, N.; Behnke, T.; Resch-Genger, U. Determination of the critical micelle concentration of neutral and ionic surfactants with fluorometry, conductometry, and surface tension—A method comparison. J. Fluoresc. 2018, 28, 465–476.
- Puvvada, S.; Blankschtein, D. Molecular-thermodynamic approach to predict micellization, phase behavior and phase separation of micellar solutions. I. Application to nonionic surfactants. J. Chem. Phys. 1990, 92, 3710–3724.
- Nagarajan, R.; Ruckenstein, E. Theory of surfactant self-assembly: A predictive molecular thermodynamic approach. Langmuir 1991, 7, 2934–2969.
- Khoshnood, A.; Lukanov, B.; Firoozabadi, A. Temperature effect on micelle formation: Molecular thermodynamic model revisited. Langmuir 2016, 32, 2175–2183.
- Glasser, L. Volume-based thermodynamics of organic liquids: Surface tension and the Eötvös equation. J. Chem. Thermodyn. 2021, 157, 106391.
- Floros, D.J.; Jensen, P.R.; Dorrestein, P.C.; Koyama, N. A metabolomics guided exploration of marine natural product chemical space. Metabolomics 2016, 12, 145.
- de Faria Poloni, J.; Bonatto, D. Systems Chemo-Biology and Transcriptomic Meta-Analysis Reveal the Molecular Roles of Bioactive Lipids in Cardiomyocyte Differentiation. J. Cell. Biochem. 2015, 116, 2018–2031.
- Feltes, B.C.; Poloni, J.d.F.; Notari, D.L.; Bonatto, D. Toxicological effects of the different substances in tobacco smoke on human embryonic development by a systems chemo-biology approach. PLoS ONE 2013, 8, e61743.
- Feltes, B.C.; de Faria Poloni, J.; Nunes, I.J.G.; Bonatto, D. Fetal alcohol syndrome, chemo-biology and OMICS: Ethanol effects on vitamin metabolism during neurodevelopment as measured by systems biology analysis. Omics J. Integr. Biol. 2014, 18, 344–363.
- Clements, T.; Rautenbach, M.; Ndlovu, T.; Khan, S.; Khan, W. A metabolomics and molecular networking approach to elucidate the structures of secondary metabolites produced by S. marcescens strains. Front. Chem. 2021, 9, 72.
- Zitnik, M.; Nguyen, F.; Wang, B.; Leskovec, J.; Goldenberg, A.; Hoffman, M.M. Machine learning for integrating data in biology and medicine: Principles, practice, and opportunities. Inf. Fusion 2019, 50, 71–91.
- Hudson, I.L. Data Integration Using Advances in Machine Learning in Drug Discovery and Molecular Biology. In Artificial Neural Networks; Cartwright, H., Ed.; Springer Science+Business Media: Oxford, UK, 2021; pp. 167–184.
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J.; Data, M. Data Mining: Practical Machine Learning Tools and Techniques; Morgan Kaufmann: Elsevier: Cambridge, MA, USA, 2016.
- Next-generation genomics: An integrative approach. Nat. Rev. Genet. 2010, 11, 476–486.
- Gomez-Cabrero, D.; Abugessaisa, I.; Maier, D.; Teschendorff, A.; Merkenschlager, M.; Gisel, A.; Ballestar, E.; Bongcam-Rudloff, E.; Conesa, A.; Tegnér, J. Data integration in the era of omics: Current and future challenges. BMC Syst. Biol. 2014, 8, I1.
- Cavill, R.; Jennen, D.; Kleinjans, J.; Briedé, J.J. Transcriptomic and metabolomic data integration. Briefings Bioinform. 2015, 17, 891–901.
- Tarazona, S.; Balzano-Nogueira, L.; Conesa, A. Chapter Eighteen—Multiomics Data Integration in Time Series Experiments. In Data Analysis for Omic Sciences: Methods and Applications; Comprehensive Analytical Chemistry; Jaumot, J., Bedia, C., Tauler, R., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 82, pp. 505–532.
- Padayachee, T.; Khamiakova, T.; Shkedy, Z.; Perola, M.; Salo, P.; Burzykowski, T. The Detection of Metabolite-Mediated Gene Module Co-Expression Using Multivariate Linear Models. PLoS ONE 2016, 11, e0150257.
- Nicolas, J. Molecular dynamics simulation of surfactin molecules at the water-hexane interface. Biophys. J. 2003, 85, 1377–1391.
- Gang, H.Z.; Liu, J.F.; Mu, B.Z. Molecular dynamics study of surfactin monolayer at the air/water interface. J. Phys. Chem. B 2011, 115, 12770–12777.
- Asadi, A.; Abdolmaleki, A.; Azizi-Shalbaf, S.; Gurushankar, K. Molecular Dynamics Study of Surfactin Interaction with Lipid Bilayer Membranes. Gene Cell Tissue 2021, 8, e112646.
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718.
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802.
- Eastman, P.; Swails, J.; Chodera, J.D.; McGibbon, R.T.; Zhao, Y.; Beauchamp, K.A.; Wang, L.P.; Simmonett, A.C.; Harrigan, M.P.; Stern, C.D.; et al. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 2017, 13, e1005659.
- Lemkul, J. From proteins to perturbed hamiltonians: A suite of tutorials for the gromacs-2018 molecular simulation package . Living J. Comput. Mol. Sci. 2018, 1, 5068.





