Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Carla Fernandes | -- | 4239 | 2022-07-03 16:56:58 | | | |
| 2 | Vivi Li | Meta information modification | 4239 | 2022-07-04 05:44:30 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Ribeiro, R.; Pinto, E.; Fernandes, C.; Sousa, E. Marine Cyclic Peptides. Encyclopedia. Available online: https://encyclopedia.pub/entry/24771 (accessed on 07 March 2026).
Ribeiro R, Pinto E, Fernandes C, Sousa E. Marine Cyclic Peptides. Encyclopedia. Available at: https://encyclopedia.pub/entry/24771. Accessed March 07, 2026.
Ribeiro, Ricardo, Eugénia Pinto, Carla Fernandes, Emília Sousa. "Marine Cyclic Peptides" Encyclopedia, https://encyclopedia.pub/entry/24771 (accessed March 07, 2026).
Ribeiro, R., Pinto, E., Fernandes, C., & Sousa, E. (2022, July 03). Marine Cyclic Peptides. In Encyclopedia. https://encyclopedia.pub/entry/24771
Ribeiro, Ricardo, et al. "Marine Cyclic Peptides." Encyclopedia. Web. 03 July, 2022.
Copy Citation
Oceans are a rich source of structurally unique bioactive compounds from the perspective of potential therapeutic agents. Marine peptides are a particularly interesting group of secondary metabolites because of their chemistry and wide range of biological activities. Among them, cyclic peptides exhibit a broad spectrum of antimicrobial activities, including against bacteria, protozoa, fungi, and viruses. Moreover, there are several examples of marine cyclic peptides revealing interesting antimicrobial activities against numerous drug-resistant bacteria and fungi, making these compounds a very promising resource in the search for novel antimicrobial agents to revert multidrug-resistance.
marine peptides
cyclic peptides
cyclic depsipeptides
antimicrobial resistance
peptide synthesis
1. Introduction
Despite great advances in the pharmaceutical and medicine fields, contagious diseases induced by bacteria, fungi, viruses, and protozoa are still a significant threat to public health, as evidenced by the SARS-Cov-2 pandemic [1]. Due to the emergence of new pathogenic agents, extensive resistance, and the lack of new drugs, contagious diseases affect both developed and developing countries [2][3].
The golden age of antibacterial agents began in the 1940–1960s, and many antibiotics dating from that period are still used in therapy today. Due to the high rate of antibiotics discovery, during this period, it was believed that infectious diseases would soon be controlled in the population [4]. A line of research on antimicrobial discovery and development was identified from many natural small-molecule products that were clinically proved to have antibacterial activity. Among these small molecules, penicillins, cephalosporins, macrolides, glycopeptides, tetracyclines and aminoglycosides stand out. On the other hand, another line of research was found from the structures of the chemical dye industry, leading to the discovery of aromatic sulfa compounds with antibiotic activity [5]. In 1960, the fluoroquinolones emerged, which are the second example of synthetic antibiotics used in therapy. Later, in the 2000s, the first generation of oxazolidinone linezolid, a synthetic derivative structurally different from the previous ones, was approved in the USA [6]. In addition, new generations of cephalosporins, macrolides, fluoroquinolones, and tetracyclines appeared with significant therapeutic use. It is important to highlight that the development of new synthetic methodologies has allowed the synthesis of pentacyclines, derived from tetracyclines, which may be considered as a fourth generation of this class of antibiotics [7].
Regarding the treatment of systemic mycoses, such as candidiasis, aspergillosis, and cryptococcosis, antifungals can be organized into four classes—polyenes, azoles, flucytosine, and echinocandins—in which they are distinguished by the mode of formulation, bioavailability, pharmacological interactions, adverse effects, and mechanism of action [8][9]. Although commensals in humans, Candida species are a cause of various infections in susceptible patients, including elderly, hospitalized, and immunosuppressed patients. Invasive Candida infection is one of the most common fungal infections globally [10]. Less common, but responsible for greater treatment concerns, are systemic infections caused by fungi of the genera Aspergillus, Fusarium and Scedosporium, considering their susceptibility profile to the available antifungals [11][12].
Over the course of human civilization, viral infections have caused millions of human casualties worldwide, driving the development of antiviral drugs in a pressing need [13]. As of April 2016, antiviral drugs have been approved to treat nine human infectious diseases: hepatitis B, hepatitis C, and infections caused by human immunodeficiency virus (HIV), human cytomegalovirus, herpes simplex virus (HSV), human papillomavirus, respiratory syncytial virus, varicella-zoster virus, and influenza virus [14][15]. Nevertheless, there is still no antiviral drug or vaccine for over 200 human viruses afflicting populations worldwide [14]. In addition, the current pneumonia outbreak caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was declared a pandemic by the World Health Organization on 11 March 2020 [16].
Parasitic diseases are a critical health concern with a profound impact on the global human population [17]. It was found that protozoans, such as Trypanosoma cruzi, Leishmania mexicana, Plasmodium falciparum, Giardia intestinalis, and Trichomonas vaginalis, are the major disease-causing parasites. The spread of infectious diseases is especially prevalent in underdeveloped countries characterized by tropical or temperate climate, as well as poor sanitary and hygienic conditions [18][19][20]. Parasite infections are the cause of 500 million deaths worldwide [21][22], and although there are drugs to fight parasite infections, these have drawbacks such as toxicity and the emergence of resistance [23][24].
Antimicrobial resistance (AMR) is another significant threat to public health systems all over the world [25]. Infection caused by microorganisms resistant to antimicrobial drugs leads to serious illnesses and elevated costs associated with more expensive antibiotics (when infections become resistant to first-line antimicrobials, treatment has to be switched to second- or third-line drugs, which are nearly always more expensive), specialized equipment, longer hospital stay, and isolation procedures for the patients. Societal costs include loss of productivity and death [25][26]. Every year, more than 2 million North Americans acquire infectious diseases associated with antibiotic-resistant microorganisms, resulting in 23,000 deaths [27]. In Europe, nearly 700 thousand cases of antibiotic-resistant infections develop directly into over 33,000 deaths yearly [28]. Despite a 36% increase in human use of antibiotics from 2000 to 2010 [29], approximately 20% of deaths worldwide are related to infectious diseases today [30]. Statistical models predicted that there were an estimated 4.95 million deaths associated with AMR in 2019, including 1.27 million deaths associated with bacterial AMR [3]. The six leading pathogens associated with AMR in 2019 (Escherichia coli, followed by Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa) were responsible for 3.57 million deaths [3].
AMR can occur through several mechanisms, whether intrinsic or acquired. Intrinsic resistance occurs naturally, as part of a microbial evolution process. Acquired resistance, on the other hand, is a consequence of the indiscriminate use of antimicrobials, and genetic mutations may occur, originating resistance genes that can be transferred between microbial species. AMR mechanisms fall into four main categories [31], as shown in Figure 1.
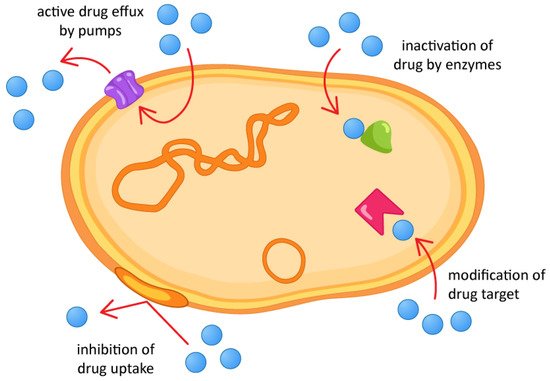
Figure 1. Antimicrobial resistance mechanisms.
Ocean water covers about 70% of the Earth’s surface and contains several potential bioactive compounds to be discovered. Marine organisms have been considered to be a promising source of numerous nutraceutical and pharmaceutical compounds [32][33]. Over the last few decades, new marine-derived compounds have been considered not only as lead compounds for drug discovery, but also as bioactive agents in pharmaceutical research, possessing antifungal, antibacterial, cytotoxic and anti-inflammatory properties, among others [34][35][36][37].
Specific chemical and physical properties, such as water salt concentrations, pressure, temperature (including extreme), light penetration, ocean currents, oxygen concentrations, and radiation exposure characterize different underwater habitats (environment) of marine organisms [38][39]. Due to this extreme environment, marine organisms are forced to produce a chemical diversity of bioactive compounds that are considered essential for the discovery and development of new agents for the treatment and prevention of various fungal, bacterial, viral, and protozoal infections [35][40][41][42][43][44][45].
In particular, several peptides have been isolated from marine sources and demonstrated to be promising drug candidates based on their significance of the bioregulatory role and unique molecular mechanisms of action [46][47]. Researchers' group also described the isolation and stereochemical analysis of marine peptides [48][49]. Compared to small-molecule drugs, peptides are highly selective and efficacious and, at the same time, relatively safe and well tolerated [50]. The high degree of selectivity in their interactions is the result of millions of years of evolutionary selection for complementary shapes and sizes from among a large array of structural and functional diversity [47].
Despite the applicability of peptides and proteins in medicine, it has been limited by their high manufacturing costs, the low bioactivity of peptides when administered orally, and their low membrane permeability, low systemic stability, and high clearance rates [51]. The low stability under physiological conditions is one of the main obstacles to the therapeutic use of linear peptides, as they easily lose their biological activity because they are rapidly cleaved by enzymes in vivo [52][53]. To overcome this obstacle, diverse peptide modifications have been proposed [54][55][56][57]. Linear peptide cyclization has recently been considered one of the most promising approaches, due to several advantages in surpassing both pharmacokinetic and pharmacodynamic limitations. A cyclic structure reduces the conformational freedom for each constituent within the cycle and forces the molecule into a more rigid secondary structure [58]. The increase in rigidity is an advantage that is translated into a decrease in the entropic term of Gibbs energy, improving binding affinities higher than some natural ligands to a biotarget [59][60][61][62]. Common motifs in the formation of proteins and polypeptides, β-turn, are other advantages of cyclization, as it is believed that this improves binding affinity [63][64][65]. Cyclization also allows the elimination of charged terminals at the ends of the structure in cyclic peptides, which may increase membrane permeability [66], although the peptide cross of the membrane does not improve just because the peptide is cyclized, but due to its structural features [67]. Another advantage of cyclization is becoming less prone to hydrolysis, as it decreases the exposure of the amino and carboxyl termini to exopeptidases [68], decreasing off-target side effects [69], thus leading to substantially enhanced metabolic stability and specificity [70][71][72][73]. Furthermore, in terms of chemical synthesis, cyclic peptides are significantly smaller when compared to proteins, and therefore, lower manufacturing costs are needed [74]. Actually, cyclic peptides are polypeptide chains that contain a circular sequence of bonds, which can be formed through a bond between the amino and carboxyl termini of the peptide with an amide bond, or other chemically stable bonds such as lactone, ether, thioether, and disulfide, among others. The formation of the amide bond between the amino and carboxylic terminals results in the formation of a head-to-tail (or N-to-C) cyclic peptide. Many cyclic peptides with this kind of formation (N-to-C) exist in nature [75][76][77][78][79].
Depending on lipophilicity, the type of bonds between amino acids, and the number of amino acids, cyclic peptides can have different classifications, as either cyclic lipopeptides, cyclic glycopeptides, or cyclic depsipeptides. Cyclic lipopeptides are cyclic peptides acylated by a lipid, usually a fatty acid (FA) side chain. These compounds are produced only in bacteria and fungi of various habitats during cultivation on carbon and nitrogen sources [80]. Cyclic glycopeptides contain carbohydrate moieties covalently attached to the side chains of amino acid residues [81]. Cyclic depsipeptides are cyclic peptides in which amide groups are replaced by corresponding lactone bonds due to the presence of a hydroxy-carboxylic acid in the peptide structure by cyclization to the hydroxyl of serine or threonine side chains [82]. Modification of the amide bond to an ester increases the lipophilicity that may subsequently improve cell permeability. Depsipeptides have also been used to demonstrate the importance of the hydrogen bonds that are formed by amide bonds in natural peptides [79]. These peptides sometimes display additional chemical modifications, including unusual amino acid residues in their structures. Cyclic depsipeptides contain at least one ring formed only through peptide or ester links—derived from hydroxy carboxylic acids [83]. Cyclic peptides and cyclic depsipeptides can be named as cyclic tri-, tetra-, penta-, hexa-, hepta-, octa-, nona-, deca-, unde-, dodeca, and tri-decapeptides, depending on the number of amino acids present [84].
Depsipeptides are also called peptolides or nonribosomal peptides (NRP), being biosynthesized by non-ribosomal peptide synthetases (NRPS) in combination with either polyketide synthase (PKS), or FA synthase enzyme systems, and are often found in marine organisms such as bacteria, tunicates, mollusks, and sponges, among others [76][77][78]. NRPS are multifunctional proteins that synthesize peptide natural products independent of the mRNA ribosomal machinery and employ a modular architecture wherein each module is responsible for the incorporation of one amino acid into the final peptide product [85][86]. NRP can be linear, cyclic, or branched peptides, and usually contain fewer than 20 amino acid residues and are often modified by chemical processes such as acylation, glycosylation, and others. Each module functions as a building block responsible for the incorporation and modification of an amino acid, especially D-amino acids, so that the order and quantity of modules in an NRPS determine the amino acid sequence of the synthesized peptide [87].
The section of the NRPS enzyme that specifically incorporates an amino acid into the peptide chain is defined as a module, and the modules, in turn, can be divided into domains, which catalyze the individual steps of non-ribosomal peptide synthesis. Each standard elongation module consists of three domains: a condensation domain (C), an adenylation domain (A), and a peptidyl transporter protein (PCP), organized as C-A-PCP [87] (Figure 2). The PCP domain is also frequently referred to as the T domain, and the holoform of this domain is a substrate for thioester (“thiolation”) formation [88][89][90][91][92]. In addition to these main domains, there are others involved in modifying an NRP: the E domain (molecule epimerization); the Cy domain (cyclization of the forming molecule); the MT domain (methylation reactions); the R domain (reduction reactions), and the Ox domain (oxidation reactions) [93].
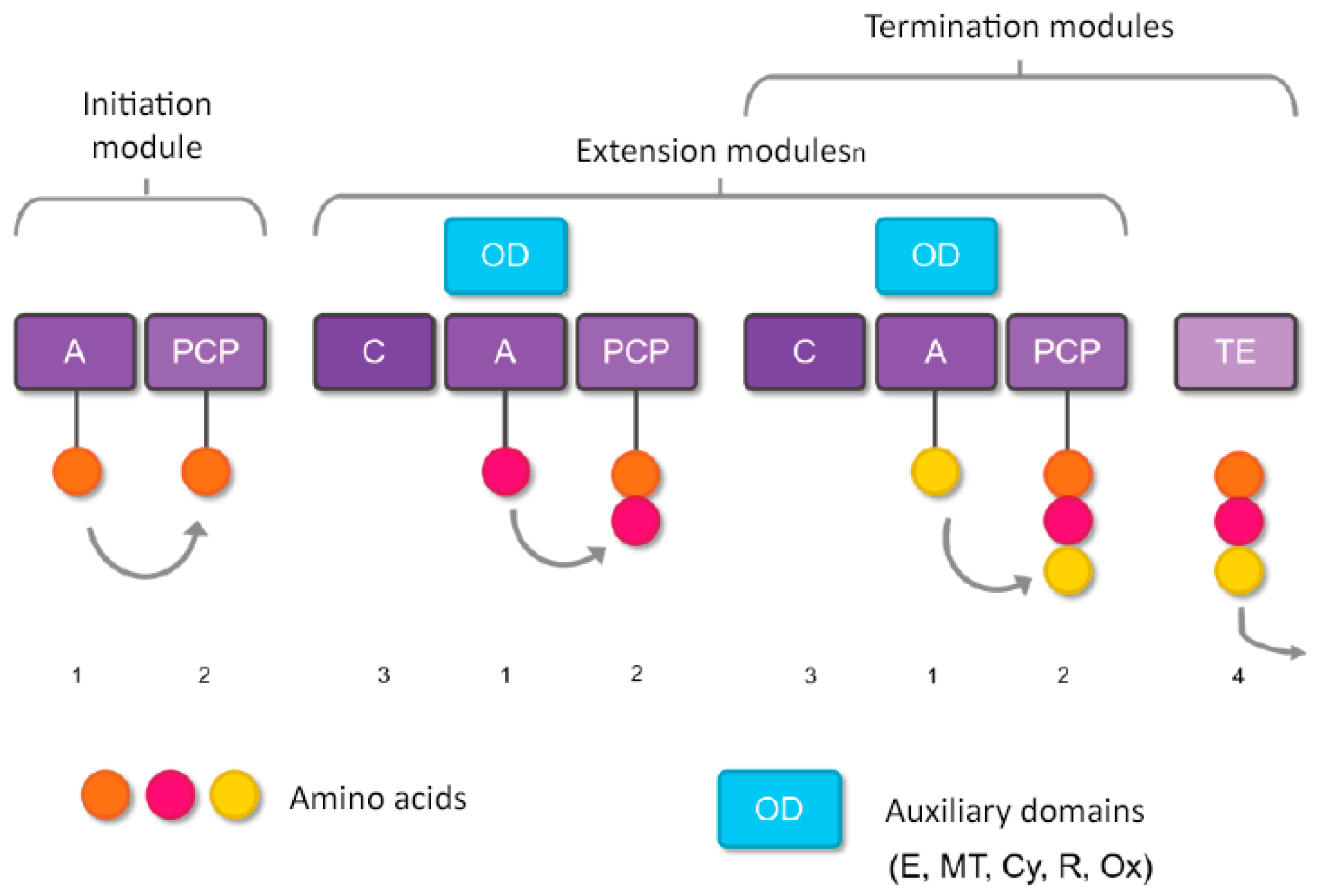
Figure 2. Basic steps of nonribosomal peptide synthesis. (1) Domain A selects the amino acid to be incorporated and transfers it to the PCP domain (2), where a thioester bond is formed. (3) Domain C forms the peptide bond between the amino acid present in the PCP domain of the same module and the intermediate peptidyl linked to the PCP domain of the previous module (that is, it catalyzes the link between amino acids of adjacent modules). (4) If no additional domains are present that modify the molecule during formation, the TE domain releases the formed peptide. However, if additional domains are present (such as E, MT, Cy, or Ox), the molecule is modified before being released by the TE domain.
The terminating domain, the thioesterase (TE) domain, normally releases the peptide by hydrolysis or cyclization [94], while reductase (R) domains catalyze release by converting the thioester link to an aldehyde [95], or alcohol [96], specialized C domains [97], catalyze cyclization [98], and amide bond formation through small molecule acceptance [97][99][100][101].
2. Marine Cyclic Peptides with Antimicrobial Activities
2.1. Sponge-Produced Cyclic Peptides
Sponges are diversified organisms distributed extensively on shores and deep in the ocean [102]. In terms of chemical diversity, an exceptionally prolific group of sponges include the lithistid sponges, the prominence of which may be due to the biosynthetic capacity of the microorganisms that host them [103][104][105]. The metabolites of lithistid sponges, which include the genera Theonella, Discodermia, Aciculites, Microscleroderma, and Callipelta, are among the most diverse found in any order of sponges and have often been attributed to symbiotic microorganisms (such as proteobacteria “Candidatus Entotheonella palauensis”) [104], that contains four distinct cell populations: sponge cells, unicellular heterotrophic bacteria, unicellular cyanobacteria, and filamentous heterotrophic bacteria. In particular, many cyclic peptides and peptide lactones have been reported to be able to be obtained from Theonella sponges, and their structural features, including unusual amino acids or D-amino acids, suggest that they perhaps originated from symbiotic microorganisms [106][107]. In this section, 63 cyclic peptides isolated from sponges (1–63) are described (Table 1 and Figure 3). Among these, 14 cyclic peptides have been found to have antibacterial activities, as well as 19 with antifungal, one with parasitic, and 22 with antiviral activities. The most relevant antimicrobial cyclic peptides are highlighted due to their unusual structural characteristics or advanced investigations, potency, and in vivo experiments.
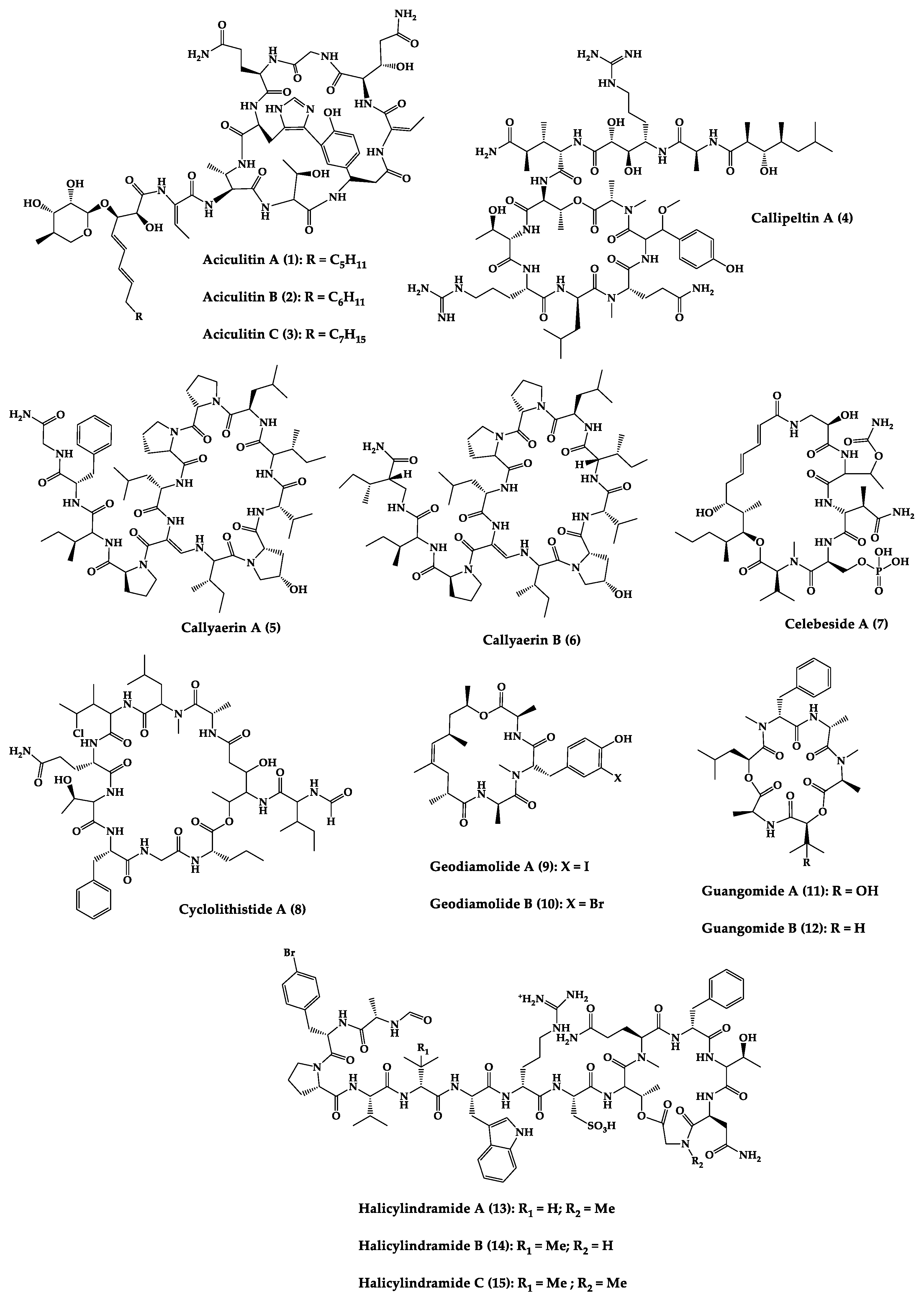
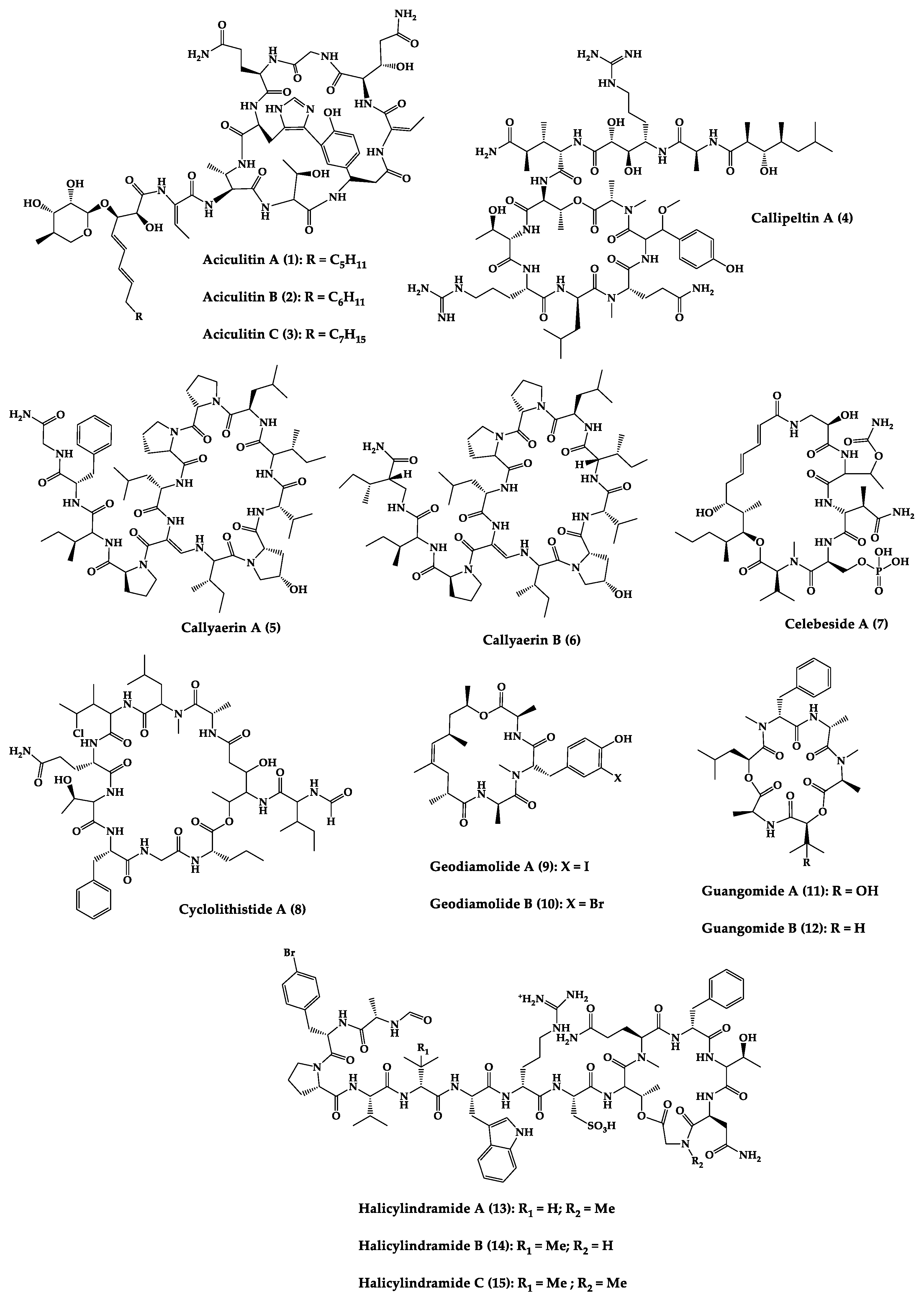
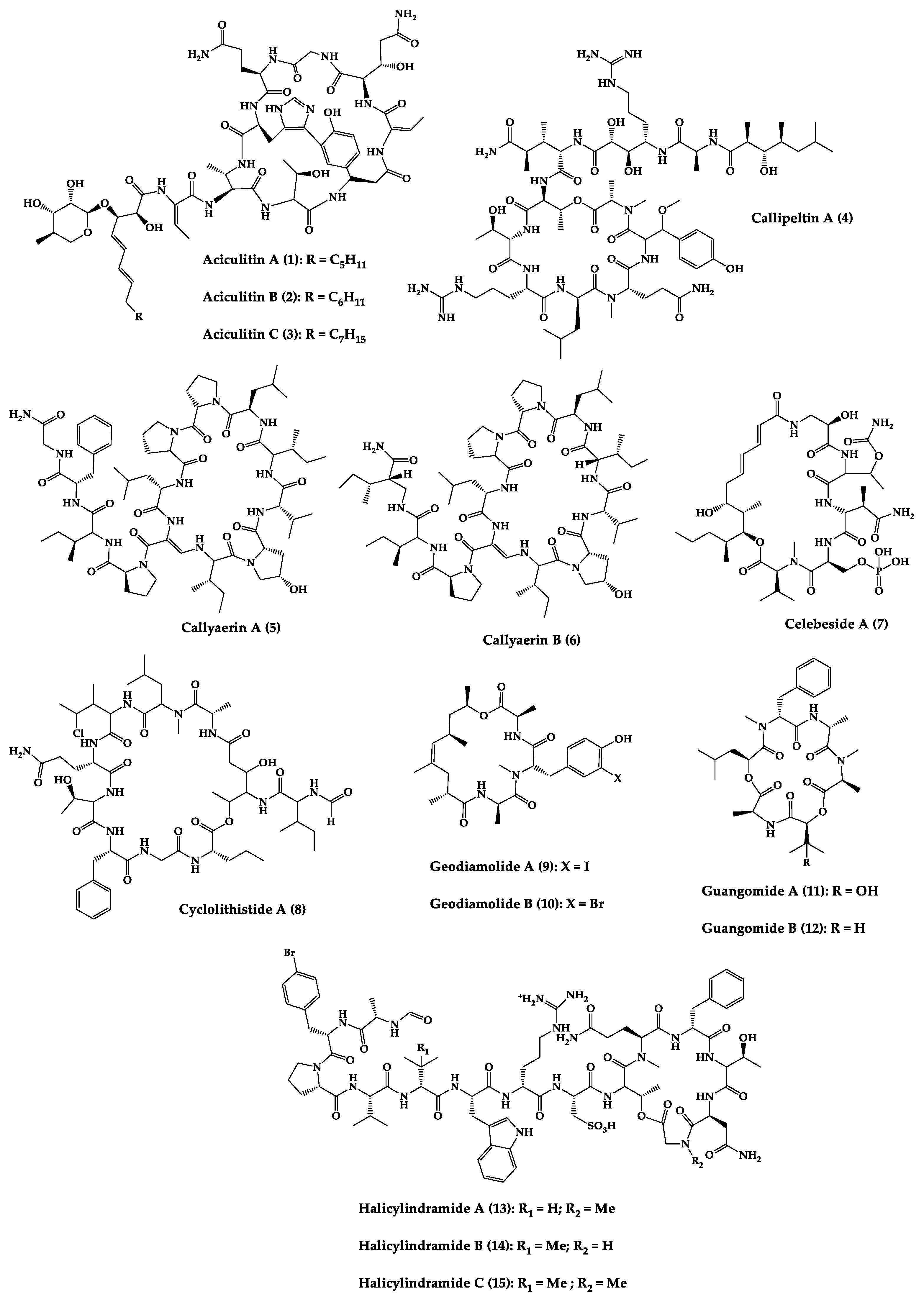
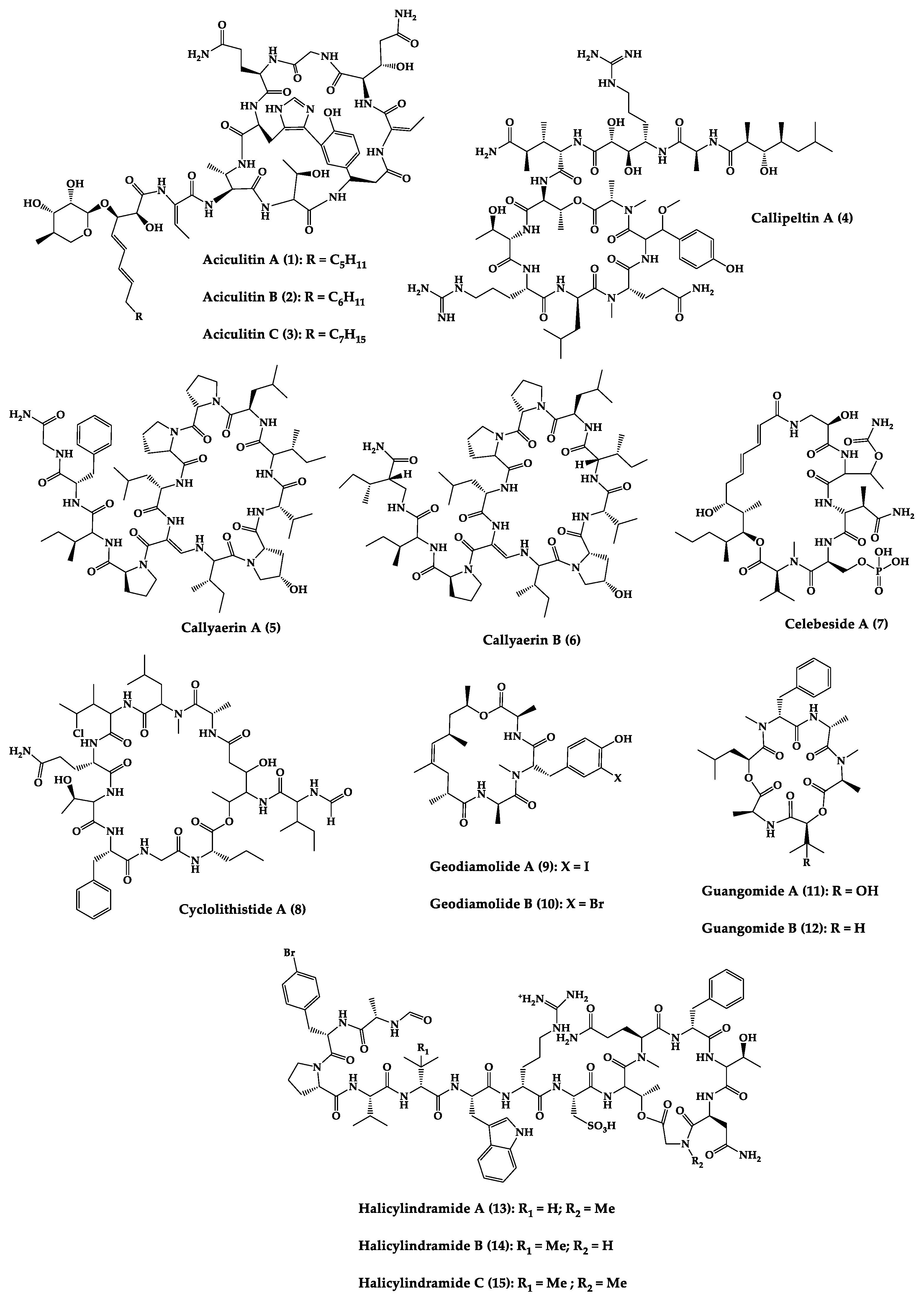
Figure 3. Chemical structures of cyclic peptides from sponges (1–63).
Halicylindramides A–C (13–15) were isolated from the Japanese marine sponge Halichondria cylindruta and are cyclic tetradecapeptides with the N-terminus blocked by a formyl group and the C-terminus lactonized with a threonine residue. Compounds 13–15 demonstrated in vitro antifungal activity against Mortierella ramanniana at 7.5 µg/disk. Interestingly, it was found that the macrocyclic structure of compounds 13–15 is essential for their cytotoxic and antifungal activities [108].
An anti-HIV undecadepsipeptide, designated homophymine A (16), isolated from a New Caledonian collection of the marine sponge Homophymia sp, contains 11 amino acid residues and an amide-linked 3-hydroxy-2,4,6-trimethyloctanoic acid moiety. This undecadepsipeptide exhibited in vitro cytoprotective activity against HIV-1 infection, demonstrating a half-maximal inhibitory concentration (IC50) of 75 nM [109] in a cell-based XTT assay.
Jasplakinolide (21), also named jaspamide, is a 19-membered macrocyclic depsipeptide isolated from the soft-bodied sponge Jaspis species collected off the shore of the island of Benga, Fiji, and shows selective in vitro antimicrobial activity with a minimum inhibitory concentration (MIC) greater than 25 µg/mL against Candida albicans. The in vivo topical activity of a 2% solution of 21 against a Candida vaginal infection in mice was similar to that of miconazole nitrate (MIC = 6.2 µg/mL) [110][111]. This macrocyclic (21) exhibited insecticidal activity against Heliothis virescens, with a lethal concentration in 50% of the population (LC50) of 4 ppm. It was also toxic to the nematode Nippo-Strongylus brasiliensis with a lethal dose in 50% of the population (LD50) of 1 µg/mL [112].
Koshikamides F (22) and H (23) are 17-residue cyclic heptadecadepsipeptides containing a 10-residue macrolactone, isolated from deep-water Palauan collections of T. swinhoei and T. cupola. Linear koshikamides (data not shown) failed to inhibit HIV entry, while the cyclic peptides 22 and 23 inhibited HIV entry with IC50 values of 2.3 and 5.5 μM, respectively. Thus, the presence of the exocyclic olefin and its associated conformation appear to enhance activity relative to the hydroxypyrrolidone version, suggesting the ten-residue lactone ring is important for inhibition of HIV-1 entry. Lastly, the slightly more potent anti-HIV activity of 22 and 23 may be due to the distinct conformation of the macrolactone brought about by the presence of the unsaturated pyrrolidinone residue 2-(3-amino-5-oxopyrrolidin-2-ylidene)propanoic acid [113].
Microspinosamide (38), a cyclic tridecadepsipeptide, incorporates numerous uncommon amino acids, and it was the first naturally occurring peptide described to contain a β-hydroxy-p-bromophenylalanine residue. Microspinosamide (38) inhibited the cytopathic effect of HIV-1 infection in an XTT-based in vitro assay with an effective concentration in 50% population (EC50) value of approximately 0.2 µg/mL [114].
Four cyclic glycodepsipeptides have been isolated from the marine sponge Siliquariaspongia mirabilis, namely mirabamides A–D (39–42), which contain 4-chlorohomoproline in 39, 40 and 41, and an unusual glycosylated amino acid, β-methoxytyrosine 4′-O-R-L-rhamnopyranoside, along with a N-terminal aliphatic hydroxy acid. Mirabamide A (39) was demonstrated in vitro to inhibit HIV-1 in neutralization and fusion assays, with IC50 values between 40 nM and 140 nM, while mirabamides C (41) and D (42) presented IC50 values of 140 nM–1.3 µM and 190 nM–3.9 µM, respectively. These results indicate that these peptides can act in the early stages of HIV-1 entry. Additionally, mirabamides E–H (43–46), which were isolated from the sponge Stelletta clavosa, collected from the Torres Strait, demonstrated in vitro HIV-1 inhibition in a neutralization assay, with IC50 values of 121 nM, 62 nM, 68 nM, and 41 nM, respectively. Some interesting structure–activity relationships (SAR) emerged by comparing the HIV inhibitory activities of mirabamides E–H (43–46) with those previously determined for mirabamides A–D (39–42) [115]. The primary feature that distinguishes compounds 43–46 from 39–42 is the presence of 2-amino-2-butenoic acid rather than threonine, and this change was found to improve activity (evidenced by the 2-fold increase in potency of 45 compared to 41). In general, increasing hydrophobicity in the side chain, but not including 2,3-diaminobutanoic acid (polar headgroup), improved potency. A potential model that may account for this tendency involves the inclusion of the hydrophobic tail into the plasma membrane by the presence of the polar headgroup (such as 2-amino-2-butenoic acid) [116]. The role of the rhamnose residue in potency is less clear. For 43–46, the absence of rhamnose is correlated with improved activity (∼2-fold) in neutralization assays, whereas the absence of rhamnose was associated with an increase and a decrease in activity for mirabamide C (41) vs. A (39) and mirabamide D (42) vs. papuamide A (51), respectively, in HIV-1 envelope-mediated fusion assays [115].
Another interesting example is neamphamide A (48), an HIV-inhibitory cyclic undecadepsipeptide, isolated from a Papua New Guinean collection of the marine sponge Neamphius huxleyi, containing 11 amino acid residues and an amide-linked 3-hydroxy-2,4,6-trimethylheptanoic acid moiety. The anti-HIV activity of 48 was evaluated in an XTT-based cell viability assay using the human T-cell line CEMSS infected with HIV-1RF [117]. After a 6-day incubation period, compound 48 effectively inhibited the cytopathic effect of HIV-1 infection with an EC50 = 28 nM [118].
Neamphamide B (49), a cyclic undecadepsipeptide, isolated from a marine sponge of Neamphius sp. collected at Okinawa, Japan, consists of uncommon amino acid residues (11 amino acid residues and an amide-linked 3-hydroxy-2,4,6-trimethylheptanoic acid moiety) and N-terminal aliphatic hydroxyl acid. The peptide 49 showed potent anti-mycobacterial activity against Mycobacterium smegmatis under standard aerobic growth conditions, as well as dormancy inducing hypoxic conditions with MIC of 1.56 µg/mL. Compound 49 was also effective against Mycobacterium bovis with MIC in the ranging of 6.25–12.5 µg/mL [119].
Papuamide A (51) and B (52) are cyclic depsipeptides isolated from bacteria symbiosis sponges Theonella mirabilis and Theonella swinhoei that exhibit a concentration-dependent increase in human T-lymphoblastoid cellular viability, indicating an inhibition of productive infection relative to control cultures, with an EC50 of 3.6 ng/mL. The HIV-inhibitory and cytotoxic activities of 52 in the same assay were virtually identical to those observed for 51 [120].
Theonellamide G (58) is a bicyclic glycodepsipeptide collected from bacteria symbiosis red sea sponge Theonella swinhoei that showed in vitro antifungal activity towards wild and amphotericin B-resistant strains of C. albicans with IC50 of 4.49 μM and 2.0 μM, respectively, compared to 1.48 μM for the positive antifungal control amphotericin-B against the wild type [121].
Theopapuamide A–C (61–63) are cyclic undecadepsipeptides isolated from bacteria symbiosis sponges Theonella swinhoei and Siliquariaspongia mirabilis, which contain several unusual amino acid residues, where the occurrence of α-methoxyasparagine and 4-amino-5-methyl-2,3,5-trihydroxyhexanoic acid is unprecedented in natural peptides. The compounds 61–63 inhibited the in vitro growth of wild-type and amphotericin B-resistant wild-type strain of C. albicans at loadings of 1–5 µg/disk, displaying zones of growth inhibition of 8 mm [122]. All theopapuamides, which lack a β-methyltyrosine residue, were inactive [115]; however, further studies demonstrated that theopapuamide B (59) was active in the neutralization assay [123] with an IC50 of 0.8 μg/mL, in an in vitro single-round HIV-1 infectivity assay against viruses pseudo-typed with HIV-1 SF162 envelope [123]. Ratnayake et al. [122] suggested that the β-methyltyrosine residue was critical for the anti-HIV activity.
According to the overall results, some remarks can be inferred. The antiviral activity results reported for mirabamide A (39) and papuamide A (51), which both contain a β-methoxytyrosine, may be justified by the fact that this residue imparts a specific conformation required for binding to target protein involved in HIV-1 entry [115]. In the case of homophymine A (16), in which the β-methoxytyrosine is replaced by an O-methyl threonine (a smaller portion of moiety and more polar), the hypothesis that β-methoxytyrosine is essential for antiviral activity is ruled out [109].
Table 1. Antimicrobial cyclic peptides from marine sponges.
| Compound | Structure | Source | Antimicrobial Activity | Synthesis | References |
|---|---|---|---|---|---|
| Aciculitins A-C (1–3) | Bicyclic octa-peptides | Aciculites orientalis | C. albicans (2.5 µg/disk, standard disk assay) | Semi-synthesis | [124][125] |
| Callipeltin A (4) | Cyclic deca-depsipeptide | Callipelta sp. | HIV-1 infection inhibition (CD50 = 0.29 µg/mL, ED50 = 0.01 µg/mL), C. albicans (100 µg/disk) | Total synthesis of analogues | [126][127][128][129][130] |
| Callyaerins A (5) and B (6) | Cyclic undeca-peptides | Callyspongia aerizusa | IC90: M. tuberculosis (2 μM and 5 μM, respectively), isoniazide (0.625 μM) | Total synthesis | [131][132] |
| Celebeside A (7) | Cyclic penta-depsipeptide | Siliquaria-spongia mirabilis | IC50: Neutralized HIV-1 (1.9 µg/mL) | - | [123] |
| Cyclolithistide A (8) | Cyclic deca-despipeptide | Bacteria symbiosis Theonella swinhoei | C. albicans (20 µg/disk) | - | [133] |
| Geodiamolides A (9) and B (10) | Cyclic depsipeptides | Geodia sp. | MIC: C. albicans (31.3 µg/mL) | Total synthesis | [134][135][136] |
| Guangomides A (11) and B (12) | Cyclic tetra-depsipeptides | Unidentifiable sponge derived fungus | MIC: S. epidermidis (100 µg/mL), E. durans (100 µg/mL) |
- | [137] |
| Halicylindramides A-C (13–15) |
Cyclic tetra-decapeptides | Halichondria cylindruta | M. ramanniana (7.5 µg/disk) | Total synthesis and analogues | [108][138][139] |
| Homophymine A (16) | Cyclic undeca-depsipeptide | Homophymia sp. | IC50: HIV-1 infection cytoprotective (75 nM) | Semi-synthesis | [109][140][141] |
| Hymenamides A (17), B (18), C (19), and E (20) | Cyclic hepta-peptides | Hymeniacidon sp. | MIC: C. albicans (33–66 µg/mL), C. neoformans (33–133 µg/mL) | Total synthesis and analogues | [142][143][144] |
| Jasplakinolide (or jaspamide) (21) | Cyclic depsipeptide | Jaspis sp. | H. virescens (LC50 = 4 ppm), N. brasiliensis (LD50 < 1 µg/mL), C. albicans (MIC > 25 µg/mL), in vivo murine vaginal C. albicans infection (2% jasplakinolide was equivalent in efficacy to administration of miconazole nitrate at 2%) |
Total synthesis and analogues | [110][111][112][145][146][147] |
| Koshikamides F (22) and H (23) | Cyclic heptadeca-peptides | Theonella swinhoei and T. cupola | IC50: HIV-1 neutralization (2.3–5.5 µM) | - | [45][113] |
| Microcionamides A (24) and B (25) | Cyclic hexapeptides | Clathria abietina | MIC: M. tuberculosis (5.7 µM) |
- | [148] |
| Microsclero-dermins A–K (26–36) and anhydromicros-clerodermin C (37) |
Cyclic hexapeptides | Cyanobacteria simbiosis Microsclero-derma herdmani sp. and Theonella sp. | C. albicans (2.5–100 µg/disk, standard disk assay) |
Total synthesis and analogues | [149][150][151][152][153] |
| Microspinosamide (38) | Cyclic trideca-depsipeptide | Sidonops microspinosa | EC50: HIV-1 infection inhibition (0.2 µg/mL) |
Semi-synthesis | [114][154] |
| Mirabamides A–H (39–46) |
Cyclic glyco-depsipeptides | Siliquarias-pongia mirabilis and Stelletta clavosa | IC50: neutralized and fusion HIV-1 (40 nM–3.9 µM), B. subtilis, C. albicans (1–5 µg/disk) |
Semi-synthesis | [115][116][155] |
| Nagahamide A (47) | Cyclic hexa-depsipeptide | Theonella swinhoei | E. coli or S. aureus (50 µg/disk, inhibition zone 7 mm) |
Semi-synthesis | [156][157] |
| Neamphamide A (48) | Cyclic undeca-depsipeptide | Neamphius huxleyi | EC50: HIV-1 infection cytoprotective (28 nM) | - | [118] |
| Neamphamide B (49) | Cyclic undeca-depsipeptide | Neamphius sp. | MIC: M. smegmatis (1.56 µg/mL), M. bovis (6.2–12.5 µg/mL) |
- | [119] |
| Neosiphoniamolide A (50) | Cyclic tetra-depsipeptide | Neosiphonia suprtes | P. oryzae (IC90 = 5 ppm) H. gramineum (MIC ≤ 2 µg/mL) | - | [158] |
| Papuamides A (51) and B (52) | Cyclic depsipeptides | Bacteria symbiosis Theonella mirabilis and Theonella swinhoei | EC50: HIV-1 infection inhibition (1–74 ng/mL) |
Total synthesis and analogues | [120][155][159][160][161][162] |
| Polydiscamide A (53) | Cyclic tridecapeptide | Discodermia sp. | MIC: B. subtilis (3.1 µg/mL) | Total synthesis and analogues | [163][164] |
| Stellettapeptins A (54) and B (55) | Cyclic undecadepsi-peptides | Microorganisms symbiosis Stelletta sp. | EC50: infection of human T-lymphoblastoid cells by HIV-1RF (23 and 27 nM, respectively) | - | [165] |
| Stylissamide G (56) | Cyclic heptapeptide | Stylissa caribica | MIC: M. audouinii, T. mentagrophytes, C. albicans (6 μg/mL) | Total Synthesis | [166] |
| Theonegramide (57) | Bicyclic glycododecapeptide | Bacteria symbiosis Theonella swinhoei | C. albicans (10 µg/disk) |
- | [167] |
| Theonellamide G (58) | Bicyclic glyco-depsipeptide | Bacteria symbiosis Theonella swinhoei | IC50: Wild and amphotericin B-resistant strains of C. albicans (2.0–4.49 μM), amphotericin-B (1.48 μM) | Semi-synthesis | [121][168] |
| Theonellapeptolide congeners 1 (59) and 2 (60) | Cyclic trideca-depsipeptides | Theonella sp. | MIC: S. aureus (8.0–16 µg/mL), M. luteus (8.0 µg/mL), B. subtilis (8.0–16 µg/mL), M. smegmatis (16–66 µg/mL), T. mentagrophytes (4.0–8.0 µg/mL), A. niger (8.0–66 µg/mL) | Total synthesis and analogues | [169][170] |
| Theopapuamide A-C (61–63) | Cyclic undeca-depsipeptides | Bacteria symbiosis Theonella swinhoei and Siliquarias-pongia mirabilis | Wild type and amphotericin B-resistant strains of C. albicans (1–5 µg/disk); in vitro HIV-1 infectivity assay IC50 = 0.8 μg/mL |
- | [123] |
CD50 (median convulsant); EC50 (effective concentration in 50% of population); ED50 (effective dose in 50% of population); HIV (human immunodeficiency virus); IC50 (half maximal inhibitory concentration); IC90 (maximum inhibitory concentration in 90% population); LD50 (lethal dose in 50% population); MIC (minimum inhibitory concentration). Aspergillus niger (A. niger); Bacillus subtilis (B. subtilis); Candida albicans (C. albicans); Cryptococcus neoformans (C. neoformans); E. coli (Escherichia coli); Enterococcus durans (E. durans); Heliothis virescens (H. virescens); Helminthosporium gramineum (H. gramineum); Micrococcus luteus (M. luteus); Microsporum audouinii (M. audouinii); Mortierella ramanniana (M. ramanniana); Mycobacterium species (M. bovis, M. smegmatis, M. tuberculosis); Nippo-Strongylus brasiliensis (N. brasiliensis); Piricularia oryzae (P. oryzae); Staphylococcus species (S. aureus, S. epidermidis); Trichophyton mentagrophytes (T. mentagrophytes).
2.2. Bacteria-Produced Cyclic Peptides
A great variety of bacteria can be found in different marine habitats. Recent studies have shown that the main phyla of marine bacteria have a wide range of inhibitory activity against different types of microorganisms. Many antimicrobial substances active in a wide range of target organisms are produced by marine bacteria [171]. In this section, 53 cyclic peptides isolated from bacteria (64–116) have been reported (Figure 4). Among these, 38 cyclic peptides have been described with antibacterial activities, as well as 14 with antifungal, five with antiparasitic, and one with antiviral activities. The most relevant antimicrobial cyclic peptides are highlighted here due to their unusual structural characteristics or advanced investigations, potency, and in vivo experiments.
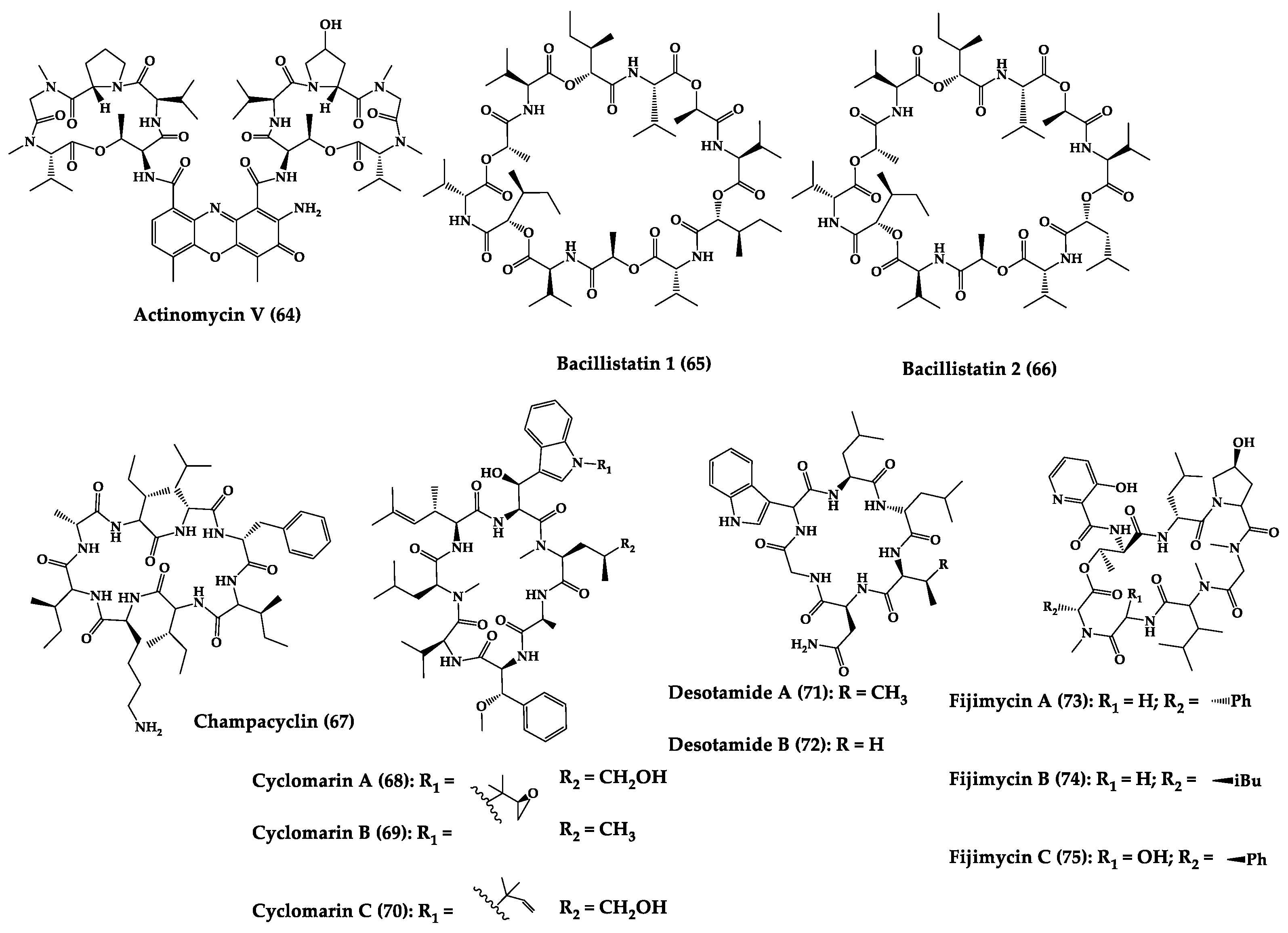
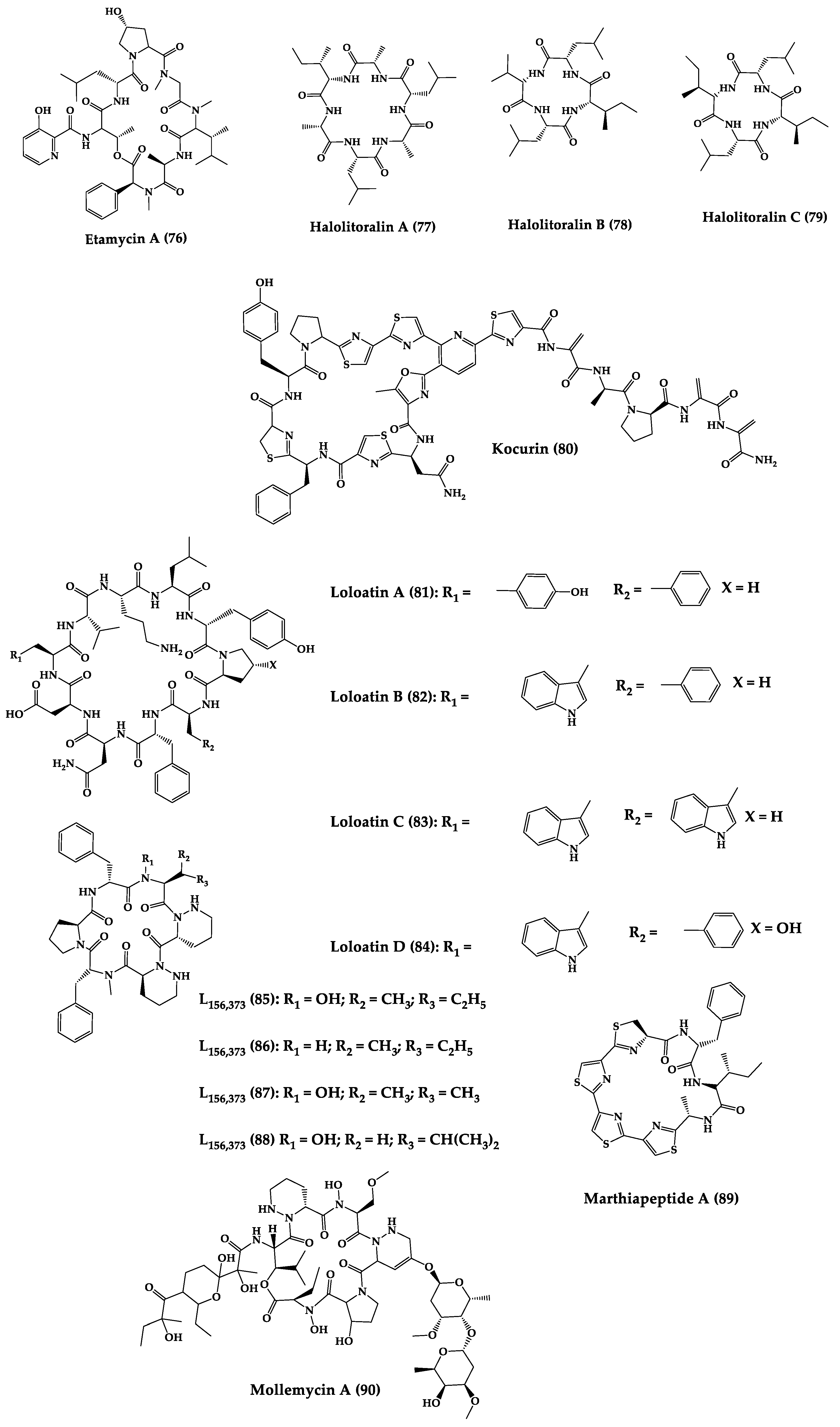
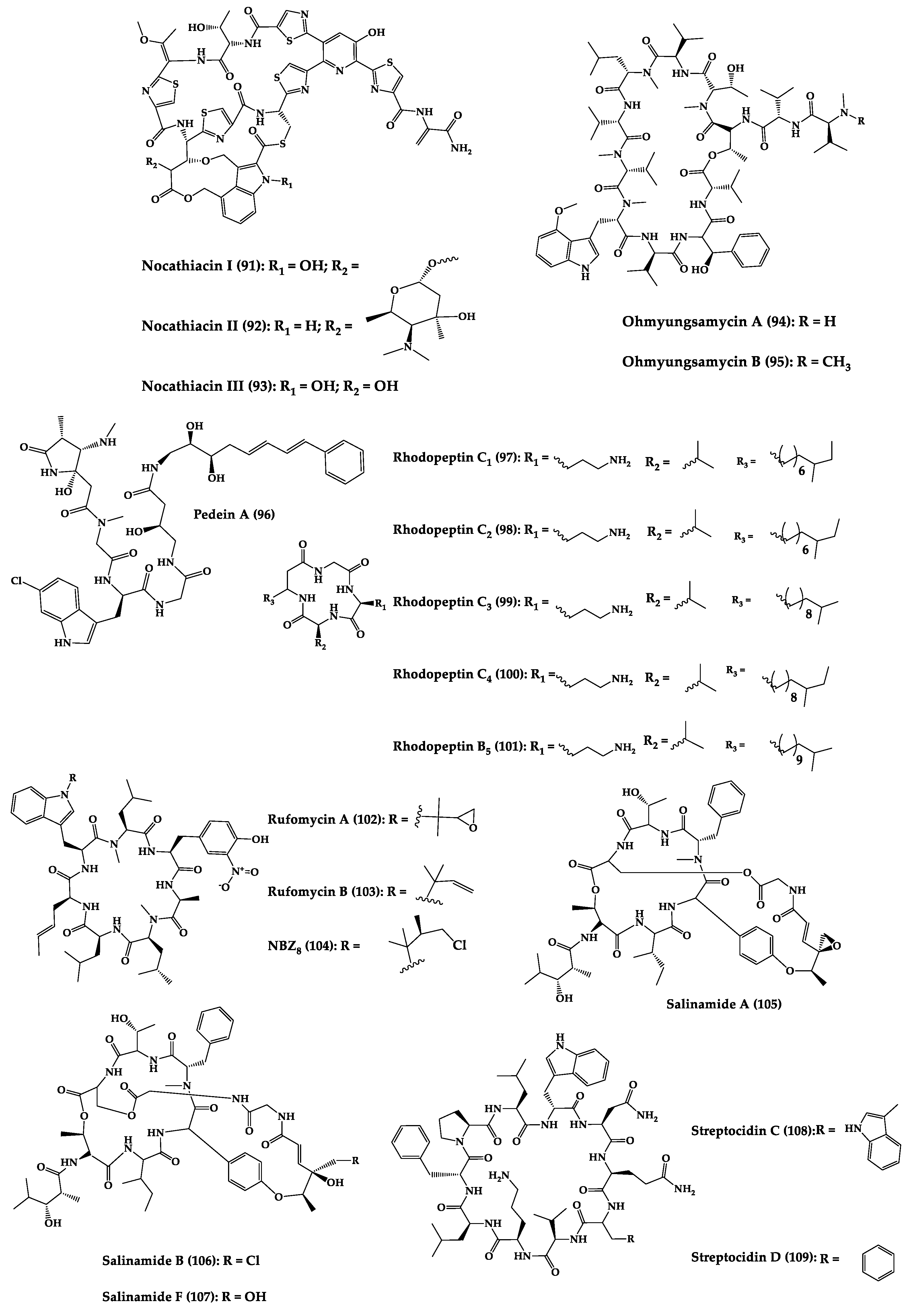
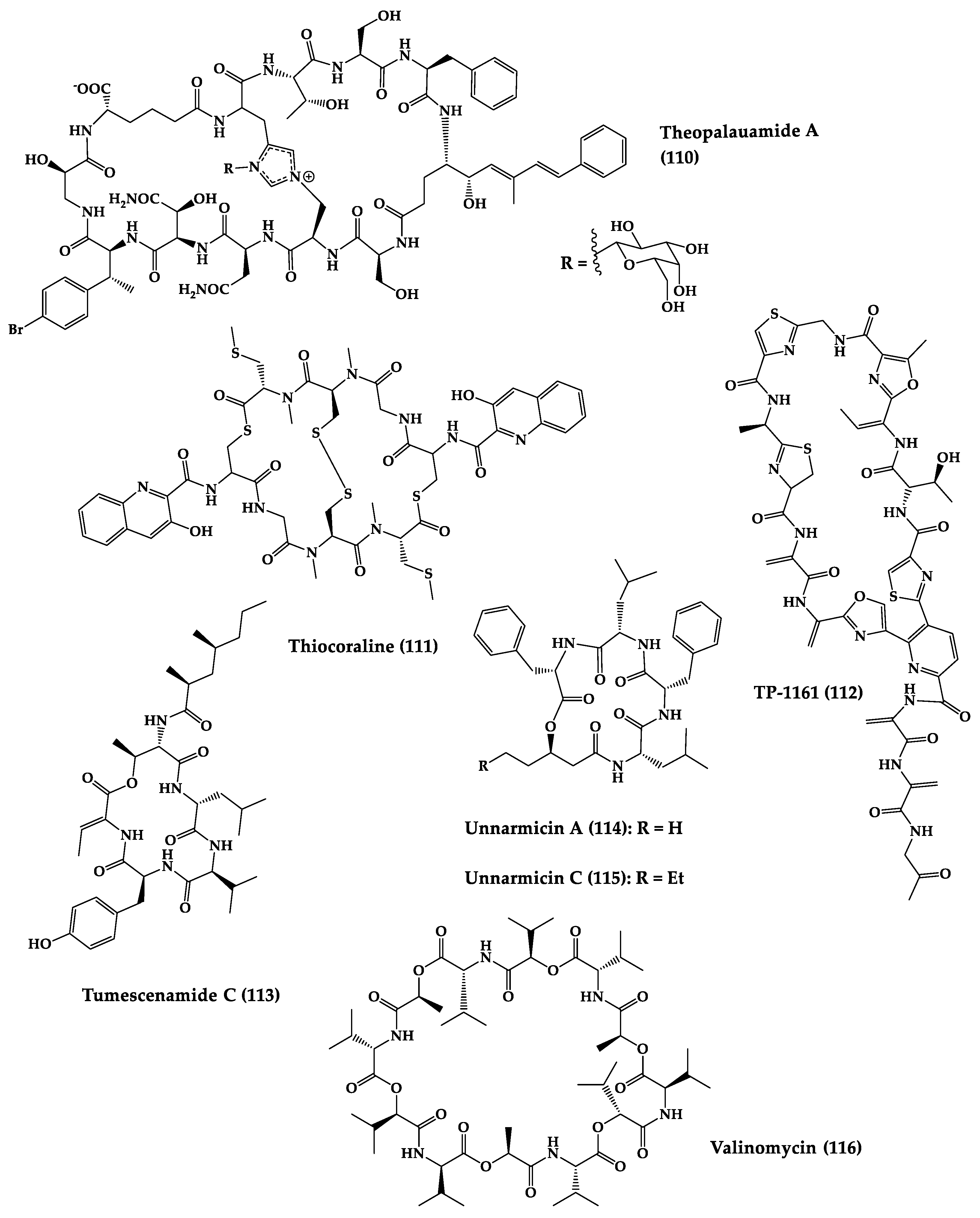
Figure 4. Chemical structures of cyclic peptides from bacteria (64–116).
References
- Loretz, B.; Oh, Y.-K.; Hudson, S.; Gu, Z.; Lehr, C.-M. Drug delivery for fighting infectious diseases: A global perspective. Drug Deliv. Transl. Res. 2021, 11, 1316–1322.
- WHO. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 12 April 2021).
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. Available online: https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(21)02724-0/fulltext (accessed on 12 April 2021).
- Livermore, D.M.; British Society for Antimicrobial Chemotherapy Working Party on The Urgent Need: Regenerating Antibacterial Drug Discovery and Development; Blaser, M.; Carrs, O.; Cassell, G.; Fishman, N.; Guidos, R.; Levy, S.; Powers, J.; Norrby, R.; et al. Discovery research: The scientific challenge of finding new antibiotics. J. Antimicrob. Chemother. 2011, 66, 1941–1944.
- Amyes, S.G. Magic Bullets, Lost Horizons: The Rise and Fall of Antibiotics, 1st ed.; CRC Press: London, UK, 2001; p. 272.
- Walsh, C.T.; Wencewicz, T.A. Prospects for new antibiotics: A molecule-centered perspective. J. Antibiot. 2014, 67, 7–22.
- Sun, C.; Hunt, D.K.; Clark, R.B.; Lofland, D.; O’Brien, W.J.; Plamondon, L.; Xiao, X.-Y. Synthesis and antibacterial activity of pentacyclines: A novel class of tetracycline analogs. J. Med. Chem. 2011, 54, 3704–3731.
- Campoy, S.; Adrio, J.L. Antifungals. Biochem. Pharmacol. 2017, 133, 86–96.
- Thompson, G.R.; Cadena, J.; Patterson, T.F. Overview of antifungal agents. Clin. Chest Med. 2009, 30, 203–215.
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163.
- Caston-Osorio, J.; Rivero, A.; Torre-Cisneros, J. Epidemiology of invasive fungal infection. Int. J. Antimicrob. Agents 2008, 32, S103–S109.
- Sobel, J.D.; Nyirjesy, P. Oteseconazole: An advance in treatment of recurrent vulvovaginal candidiasis. Future Microbiol. 2021, 16, 1453–1461.
- De Clercq, E. Recent highlights in the development of new antiviral drugs. Curr. Opin. Microbiol. 2005, 8, 552–560.
- Woolhouse, M.; Scott, F.; Hudson, Z.; Howey, R.; Chase-Topping, M. Human viruses: Discovery and emergence. Philos. Trans. R. Soc. B 2012, 367, 2864–2871.
- De Clercq, E.; Li, G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016, 29, 695–747.
- Wu, J.T.; Leung, K.; Leung, G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet 2020, 395, 689–697.
- Nash, T.E. Parasitic Diseases that Cause Seizures: Parasitic Diseases that Cause Seizures. Epilepsy Curr. 2014, 14, 29–34.
- Renslo, A.R.; McKerrow, J.H. Drug discovery and development for neglected parasitic diseases. Nat. Chem. Biol. 2006, 2, 701–710.
- Pozio, E. World distribution of Trichinella spp. infections in animals and humans. Vet. Parasitol. 2007, 149, 3–21.
- Garcia, H.H.; Gonzalez, A.E.; Gilman, R.H. Diagnosis, treatment and control of Taenia solium cysticercosis. Curr. Opin. Infect. Dis. 2003, 16, 411–419.
- Pink, R.; Hudson, A.; Mouriès, M.-A.; Bendig, M. Opportunities and challenges in antiparasitic drug discovery. Nat. Rev. Drug Discov. 2005, 4, 727–740.
- Watkins, B.M. Drugs for the control of parasitic diseases: Current status and development. Trends Parasitol. 2003, 19, 477–478.
- Upcroft, J.A.; Dunn, L.A.; Wal, T.; Tabrizi, S.; Delgadillo-Correa, M.G.; Johnson, P.J.; Garland, S.; Siba, P.; Upcroft, P. Metronidazole resistance in Trichomonas vaginalis from highland women in Papua New Guinea. Sex Health 2009, 6, 334–338.
- Anthwal, A.; Rajesh, U.C.; Rawat, M.; Kushwaha, B.; Maikhuri, J.P.; Sharma, V.L.; Gupta, G.; Rawat, D.S. Novel metronidazole–chalcone conjugates with potential to counter drug resistance in Trichomonas vaginalis. Eur. J. Med. Chem. 2014, 79, 89–94.
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318.
- Shrestha, P.; Cooper, B.S.; Coast, J.; Oppong, R.; Thuy, N.D.T.; Phodha, T.; Celhay, O.; Guerin, P.J.; Wertheim, H.; Lubell, Y. Enumerating the economic cost of antimicrobial resistance per antibiotic consumed to inform the evaluation of interventions affecting their use. Antimicrob. Resist. Infect. Control. 2018, 7, 1–9.
- Groseclose, S.L.; Buckeridge, D.L. Public health surveillance systems: Recent advances in their use and evaluation. Annu. Rev. Public Health 2017, 38, 57–79.
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66.
- Laxminarayan, R.; Matsoso, P.; Pant, S.; Brower, C.; Røttingen, J.-A.; Klugman, K.; Davies, S. Access to effective antimicrobials: A worldwide challenge. Lancet 2016, 387, 168–175.
- Martens, E.; Demain, A.L. The antibiotic resistance crisis, with a focus on the United States. J. Antibiot. 2017, 70, 520–526.
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updates 2016, 26, 43–57.
- Anjum, K.; Abbas, S.Q.; Shah, S.A.A.; Akhter, N.; Batool, S.; ul Hassan, S.S. Marine sponges as a drug treasure. Biomol. Ther. 2016, 24, 347.
- Mayer, A.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Fusetani, N. Marine pharmacology in 2009–2011: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2013, 11, 2510–2573.
- Cheung, R.C.F.; Ng, T.B.; Wong, J.H. Marine peptides: Bioactivities and applications. Mar. Drugs 2015, 13, 4006–4043.
- Mayer, A.M.; Rodríguez, A.D.; Berlinck, R.G.; Fusetani, N. Marine pharmacology in 2007–8: Marine compounds with antibacterial, anticoagulant, antifungal, anti-inflammatory, antimalarial, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous system, and other miscellaneous mechanisms of action. Comp. Biochem. Physiol. C 2011, 153, 191–222.
- Aneiros, A.; Garateix, A. Bioactive peptides from marine sources: Pharmacological properties and isolation procedures. J. Chromatogr. B 2004, 803, 41–53.
- Donia, M.; Hamann, M.T. Marine natural products and their potential applications as anti-infective agents. Lancet Infect. Dis. 2003, 3, 338–348.
- Koslow, J.A. The silent deep: The discovery, ecology, and conservation of the deep sea. Oceanography 2007, 23, 228.
- Russo, P.; Del Bufalo, A.; Fini, M. Deep sea as a source of novel-anticancer drugs: Update on discovery and preclinical/clinical evaluation in a systems medicine perspective. EXCLI J. 2015, 14, 228.
- Xu, L.; Meng, W.; Cao, C.; Wang, J.; Shan, W.; Wang, Q. Antibacterial and antifungal compounds from marine fungi. Mar. Drugs 2015, 13, 3479–3513.
- Alves, A.; Sousa, E.; Kijjoa, A.; Pinto, M. Marine-derived compounds with potential use as cosmeceuticals and nutricosmetics. Molecules 2020, 25, 2536.
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 2014, 12, 4539–4577.
- Bhatnagar, I.; Kim, S.-K. Immense essence of excellence: Marine microbial bioactive compounds. Mar. Drugs 2010, 8, 2673–2701.
- Hu, Y.; Chen, J.; Hu, G.; Yu, J.; Zhu, X.; Lin, Y.; Chen, S.; Yuan, J. Statistical research on the bioactivity of new marine natural products discovered during the 28 years from 1985 to 2012. Mar. Drugs 2015, 13, 202–221.
- Winder, P.L.; Pomponi, S.A.; Wright, A.E. Natural Products from the Lithistida: A Review of the Literature since 2000. Mar. Drugs 2011, 9, 2643–2682.
- Duray, H.; Hatfill, J.; Pellis, R. Venom peptides as human pharmaceuticals. Sci. Med. 1997, 4, 6–15.
- Wang, X.; Gong, X.; Li, P.; Lai, D.; Zhou, L. Structural diversity and biological activities of cyclic depsipeptides from fungi. Molecules 2018, 23, 169.
- May Zin, W.W.; Buttachon, S.; Dethoup, T.; Fernandes, C.; Cravo, S.; Pinto, M.M.; Gales, L.; Pereira, J.A.; Silva, A.; Sekeroglu, N. New cyclotetrapeptides and a new diketopiperzine derivative from the marine sponge-associated fungus Neosartorya glabra KUFA 0702. Mar. Drugs 2016, 14, 136.
- Prompanya, C.; Fernandes, C.; Cravo, S.; Pinto, M.M.; Dethoup, T.; Silva, A.; Kijjoa, A. A new cyclic hexapeptide and a new isocoumarin derivative from the marine sponge-associated fungus Aspergillus similanensis KUFA 0013. Mar. Drugs 2015, 13, 1432–1450.
- Cooper, B.M.; Iegre, J.; O’Donovan, D.H.; Halvarsson, M.Ö.; Spring, D.R. Peptides as a platform for targeted therapeutics for cancer: Peptide–drug conjugates (PDCs). Chem. Soc. Rev. 2021, 50, 1480–1494.
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013, 81, 136–147.
- Zompra, A.A.; Galanis, A.S.; Werbitzky, O.; Albericio, F. Manufacturing peptides as active pharmaceutical ingredients. Future Med. Chem. 2009, 1, 361–377.
- Otvos, L., Jr. Peptide-based drug design: Here and now. Methods Mol. Biol. 2008, 494, 1–8.
- Goodwin, D.; Simerska, P.; Toth, I. Peptides as therapeutics with enhanced bioactivity. Curr. Med. Chem. 2012, 19, 4451–4461.
- Ahrens, V.M.; Frank, R.; Boehnke, S.; Schütz, C.L.; Hampel, G.; Iffland, D.S.; Bings, N.H.; Hey-Hawkins, E.; Beck-Sickinger, A.G. Receptor-mediated uptake of boron-rich neuropeptide y analogues for boron neutron capture therapy. ChemMedChem 2015, 10, 164–172.
- Mason, J.M. Design and development of peptides and peptide mimetics as antagonists for therapeutic intervention. Future Med. Chem. 2010, 2, 1813–1822.
- Gentilucci, L.; De Marco, R.; Cerisoli, L. Chemical modifications designed to improve peptide stability: Incorporation of non-natural amino acids, pseudo-peptide bonds, and cyclization. Curr. Pharm. Des. 2010, 16, 3185–3203.
- Guharoy, M.; Chakrabarti, P. Secondary structure based analysis and classification of biological interfaces: Identification of binding motifs in protein–protein interactions. Bioinformatics 2007, 23, 1909–1918.
- DeLorbe, J.E.; Clements, J.H.; Whiddon, B.B.; Martin, S.F. Thermodynamic and structural effects of macrocyclic constraints in protein–ligand interactions. ACS Med. Chem. Lett. 2010, 1, 448–452.
- DeLorbe, J.E.; Clements, J.H.; Teresk, M.G.; Benfield, A.P.; Plake, H.R.; Millspaugh, L.E.; Martin, S.F. Thermodynamic and Structural Effects of Conformational Constraints in Protein–Ligand Interactions. Entropic Paradoxy Associated with Ligand Preorganization. J. Am. Chem. Soc. 2009, 131, 16758–16770.
- Tapeinou, A.; Matsoukas, M.T.; Simal, C.; Tselios, T. Review cyclic peptides on a merry-go-round; towards drug design. Pept. Sci. 2015, 104, 453–461.
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into protein–ligand interactions: Mechanisms, models, and methods. Int. J. Mol. Sci. 2016, 17, 144.
- Lee, M.S.; Gardner, B.; Kahn, M.; Nakanishi, H. The three dimensional solution structure of a constrained peptidomimetic in water and in chloroform observation of solvent induced hydrophobic cluster. FEBS Lett. 1995, 359, 113–118.
- Uma, K.; Kishore, R.; Balaram, P. Stereochemical constraints in peptide design: Analysis of the influence of a disulfide bridge and an α-aminoisobutyryl residue on the conformation of a hexapeptide. Biopolymers 1993, 33, 865–871.
- Lau, D.; Guo, L.; Liu, R.; Marik, J.; Lam, K. Peptide ligands targeting integrin α3β1 in non-small cell lung cancer. Lung Cancer 2006, 52, 291–297.
- Rezai, T.; Yu, B.; Millhauser, G.L.; Jacobson, M.P.; Lokey, R.S. Testing the conformational hypothesis of passive membrane permeability using synthetic cyclic peptide diastereomers. J. Am. Chem. Soc. 2006, 128, 2510–2511.
- Kwon, Y.-U.; Kodadek, T. Quantitative comparison of the relative cell permeability of cyclic and linear peptides. Chem. Biol. 2007, 14, 671–677.
- Hussack, G.; Hirama, T.; Ding, W.; MacKenzie, R.; Tanha, J. Engineered single-domain antibodies with high protease resistance and thermal stability. PLoS ONE 2011, 6, e28218.
- Lindgren, M.; Hällbrink, M.; Prochiantz, A.; Langel, Ü. Cell-penetrating peptides. Trends Pharmacol. Sci. 2000, 21, 99–103.
- Cini, E.; Bifulco, G.; Menchi, G.; Rodriquez, M.; Taddei, M. Synthesis of Enantiopure 7-substituted Azepane-2-carboxylic acids as templates for conformationally constrained Peptidomimetics. Eur. J. Org. Chem. 2012, 2012, 2133–2141.
- Grigoryan, G.; Reinke, A.W.; Keating, A.E. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature 2009, 458, 859–864.
- Grauer, A.; König, B. Peptidomimetics—A versatile route to biologically active compounds. Eur. J. Org. Chem. 2009, 2009, 5099–5111.
- Mandell, D.J.; Kortemme, T. Computer-aided design of functional protein interactions. Nat. Chem. Biol. 2009, 5, 797–807.
- Terrett, N. Drugs in middle space. MedChemComm 2013, 4, 474–475.
- Joo, S.H. Cyclic peptides as therapeutic agents and biochemical tools. Biomol. Ther. 2012, 20, 19.
- Gerwick, W.H.; Fenner, A.M. Drug discovery from marine microbes. Microb. Ecol. 2013, 65, 800–806.
- Rocha-Martin, J.; Harrington, C.; Dobson, A.D.; O’Gara, F. Emerging strategies and integrated systems microbiology technologies for biodiscovery of marine bioactive compounds. Mar. Drugs 2014, 12, 3516–3559.
- Kang, H.K.; Seo, C.H.; Park, Y. Marine peptides and their anti-infective activities. Mar. Drugs 2015, 13, 618–654.
- Bionda, N.; Stawikowski, M.; Stawikowska, R.; Cudic, M.; López-Vallejo, F.; Treitl, D.; Medina-Franco, J.; Cudic, P. Effects of cyclic lipodepsipeptide structural modulation on stability, antibacterial activity, and human cell toxicity. ChemMedChem 2012, 7, 871–882.
- Raaijmakers, J.M.; De Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062.
- Jensen, K.J.; Brask, J. Carbohydrates in peptide and protein design. Pept. Sci. 2005, 80, 747–761.
- Taevernier, L.; Wynendaele, E.; Gevaert, B.; De Spiegeleer, B. Chemical classification of cyclic depsipeptides. Curr. Protein Pept. Sci. 2017, 18, 425–452.
- Moss, G.; Smith, P.; Tavernier, D. Glossary of class names of organic compounds and reactivity intermediates based on structure (IUPAC Recommendations 1995). Pure Appl. Chem. 1995, 67, 1307–1375.
- Dixon, H.B.F. Nomenclature and symbolism for amino acids and peptides. Pure Appl. Chem. 1984, 56, 595–624. Available online: https://febs.onlinelibrary.wiley.com/doi/10.1111/j.1432-1033.1984.tb07877.x (accessed on 12 April 2021).
- Sieber, S.A.; Marahiel, M.A. Molecular mechanisms underlying nonribosomal peptide synthesis: Approaches to new antibiotics. Chem. Rev. 2005, 105, 715–738.
- Fischbach, M.A.; Walsh, C.T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem. Rev. 2006, 106, 3468–3496.
- Thompson, F.T.C. Biossíntese de Metabólitos Secundários. In Biotecnologia Marinha; FURG, Ed.; PPG-Mar: Rio Grande, Mexico, 2020; pp. 91–95.
- Reimer, J.M.; Haque, A.S.; Tarry, M.J.; Schmeing, T.M. Piecing together nonribosomal peptide synthesis. Curr. Opin. Struct. Biol. 2018, 49, 104–113.
- Gulick, A.M. Structural insight into the necessary conformational changes of modular nonribosomal peptide synthetases. Curr. Opin. Chem. Biol. 2016, 35, 89–96.
- Schwarzer, D.; Finking, R.; Marahiel, M.A. Nonribosomal peptides: From genes to products. Nat. Prod. Rep. 2003, 20, 275–287.
- Marahiel, M.A.; Stachelhaus, T.; Mootz, H.D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 1997, 97, 2651–2674.
- Alonzo, D.A.; Schmeing, T.M. Biosynthesis of depsipeptides, or Depsi: The peptides with varied generations. Protein Sci. 2020, 29, 2316–2347.
- Donadio, S.; Monciardini, P.; Sosio, M. Polyketide synthases and nonribosomal peptide synthetases: The emerging view from bacterial genomics. Nat. Prod. Rep. 2007, 24, 1073–1109.
- Horsman, M.E.; Hari, T.P.; Boddy, C.N. Polyketide synthase and non-ribosomal peptide synthetase thioesterase selectivity: Logic gate or a victim of fate? Nat. Prod. Rep. 2016, 33, 183–202.
- Wilson, D.J.; Shi, C.; Teitelbaum, A.M.; Gulick, A.M.; Aldrich, C.C. Characterization of AusA: A dimodular nonribosomal peptide synthetase responsible for the production of aureusimine pyrazinones. Biochemistry 2013, 52, 926–937.
- Manavalan, B.; Murugapiran, S.K.; Lee, G.; Choi, S. Molecular modeling of the reductase domain to elucidate the reaction mechanism of reduction of peptidyl thioester into its corresponding alcohol in non-ribosomal peptide synthetases. BMC Struct. Biol. 2010, 10, 1–14.
- Bloudoff, K.; Schmeing, T.M. Structural and functional aspects of the nonribosomal peptide synthetase condensation domain superfamily: Discovery, dissection and diversity. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1587–1604.
- Zhang, J.; Liu, N.; Cacho, R.A.; Gong, Z.; Liu, Z.; Qin, W.; Tang, C.; Tang, Y.; Zhou, J. Structural basis of nonribosomal peptide macrocyclization in fungi. Nat. Chem. Biol. 2016, 12, 1001–1003.
- Bloudoff, K.; Fage, C.D.; Marahiel, M.A.; Schmeing, T.M. Structural and mutational analysis of the nonribosomal peptide synthetase heterocyclization domain provides insight into catalysis. Proc. Natl. Acad. Sci. USA 2017, 114, 95–100.
- Yuwen, L.; Zhang, F.-L.; Chen, Q.-H.; Lin, S.-J.; Zhao, Y.-L.; Li, Z.-Y. The role of aromatic L-amino acid decarboxylase in bacillamide C biosynthesis by Bacillus atrophaeus C89. Sci. Rep. 2013, 3, 1–10.
- Chang, C.-Y.; Lohman, J.R.; Huang, T.; Michalska, K.; Bigelow, L.; Rudolf, J.D.; Jedrzejczak, R.; Yan, X.; Ma, M.; Babnigg, G. Structural insights into the free-standing condensation enzyme SgcC5 catalyzing ester-bond formation in the biosynthesis of the enediyne antitumor antibiotic C-1027. Biochemistry 2018, 57, 3278–3288.
- Anand, T.P.; Chellaram, C.; Kuberan, G.; Archana, H. Bioactive peptides from marine sources-a review. Indian J. Innov. Dev. 2012, 1, 61–64.
- Bewley, C.A.; Faulkner, D.J. Lithistid sponges: Star performers or hosts to the stars. Angew. Chem. Int. Ed. 1998, 37, 2162–2178.
- Schmidt, E.; Obraztsova, A.; Davidson, S.; Faulkner, D.; Haygood, M. Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-proteobacterium,“Candidatus Entotheonella palauensis”. Mar. Biol. 2000, 136, 969–977.
- Amelia, T.S.M.; Suaberon, F.A.C.; Vad, J.; Fahmi, A.D.M.; Saludes, J.P.; Bhubalan, K. Recent Advances of Marine Sponge-Associated Microorganisms as a Source of Commercially Viable Natural Products. Mar. Biotechnol. 2022, 1–21.
- Liu, L.; Zheng, Y.-Y.; Shao, C.-L.; Wang, C.-Y. Metabolites from marine invertebrates and their symbiotic microorganisms: Molecular diversity discovery, mining, and application. Mar. Life Sci. Technol. 2019, 1, 60–94.
- Kobayashi, M.; Tanaka, J.-I.; Katori, T.; Matsuura, M.; Yamashita, M.; Kitagawa, I. Marine natural products. XXII: The absolute stereostructure of swinholide A, a potent cytotoxic dimeric macrolide from the Okinawan marine sponge Theonella swinhoei. Chem. Pharm. Bull. 1990, 38, 2409–2418.
- Li, H.-Y.; Matsunaga, S.; Fusetani, N. Halicylindramides A-C, antifungal and cytotoxic depsipeptides from the marine sponge Halichondria cylindrata. J. Med. Chem. 1995, 38, 338–343.
- Zampella, A.; Sepe, V.; Luciano, P.; Bellotta, F.; Monti, M.C.; D’Auria, M.V.; Jepsen, T.; Petek, S.; Adeline, M.-T.; Laprévôte, O. Homophymine A, an anti-HIV cyclodepsipeptide from the sponge Homophymia sp. J. Org. Chem. 2008, 73, 5319–5327.
- Zabriskie, T.M.; Klocke, J.A.; Ireland, C.M.; Marcus, A.H.; Molinski, T.F.; Faulkner, D.J.; Xu, C.; Clardy, J. Jaspamide, a modified peptide from a Jaspis sponge, with insecticidal and antifungal activity. J. Am. Chem. Soc. 1986, 108, 3123–3124.
- Scott, V.; Boehme, R.; Matthews, T. New class of antifungal agents: Jasplakinolide, a cyclodepsipeptide from the marine sponge, Jaspis species. Antimicrob. Agents Chemother. 1988, 32, 1154–1157.
- Fusetani, N.; Matsunaga, S. Bioactive sponge peptides. Chem. Rev. 1993, 93, 1793–1806.
- Plaza, A.; Bifulco, G.; Masullo, M.; Lloyd, J.R.; Keffer, J.L.; Colin, P.L.; Hooper, J.N.; Bell, L.J.; Bewley, C.A. Mutremdamide A and koshikamides C–H, peptide inhibitors of HIV-1 entry from different Theonella species. J. Org. Chem. 2010, 75, 4344–4355.
- Rashid, M.A.; Gustafson, K.R.; Cartner, L.K.; Shigematsu, N.; Pannell, L.K.; Boyd, M.R. Microspinosamide, a New HIV-Inhibitory Cyclic Depsipeptide from the Marine Sponge Sidonops microspinosa. J. Nat. Prod. 2001, 64, 117–121.
- Plaza, A.; Gustchina, E.; Baker, H.L.; Kelly, M.; Bewley, C.A. Mirabamides A–D, depsipeptides from the sponge Siliquariaspongia mirabilis that inhibit HIV-1 fusion. J. Nat. Prod. 2007, 70, 1753–1760.
- Lu, Z.; Van Wagoner, R.M.; Harper, M.K.; Baker, H.L.; Hooper, J.N.; Bewley, C.A.; Ireland, C.M. Mirabamides E–H, HIV-inhibitory depsipeptides from the sponge Stelletta clavosa. J. Nat. Prod. 2011, 74, 185–193.
- Gulakowski, R.J.; McMahon, J.B.; Staley, P.G.; Moran, R.A.; Boyd, M.R. A semiautomated multiparameter approach for anti-HIV drug screening. J. Virol. Methods 1991, 33, 87–100.
- Oku, N.; Gustafson, K.R.; Cartner, L.K.; Wilson, J.A.; Shigematsu, N.; Hess, S.; Pannell, L.K.; Boyd, M.R.; McMahon, J.B. Neamphamide A, a new HIV-inhibitory depsipeptide from the Papua New Guinea marine sponge Neamphius huxleyi. J. Nat. Prod. 2004, 67, 1407–1411.
- Yamano, Y.; Arai, M.; Kobayashi, M. Neamphamide B, new cyclic depsipeptide, as an anti-dormant mycobacterial substance from a Japanese marine sponge of Neamphius sp. Bioorg. Med. Chem. Lett. 2012, 22, 4877–4881.
- Ford, P.W.; Gustafson, K.R.; McKee, T.C.; Shigematsu, N.; Maurizi, L.K.; Pannell, L.K.; Williams, D.E.; Dilip de Silva, E.; Lassota, P.; Allen, T.M. Papuamides A–D, HIV-Inhibitory and Cytotoxic Depsipeptides from the Sponges Theonella mirabilis and Theonella swinhoei Collected in Papua New Guinea. J. Am. Chem. Soc. 1999, 121, 5899–5909.
- Youssef, D.T.; Shaala, L.A.; Mohamed, G.A.; Badr, J.M.; Bamanie, F.H.; Ibrahim, S.R. Theonellamide G, a potent antifungal and cytotoxic bicyclic glycopeptide from the Red Sea marine sponge Theonella swinhoei. Mar. Drugs 2014, 12, 1911–1923.
- Ratnayake, A.S.; Bugni, T.S.; Feng, X.; Harper, M.K.; Skalicky, J.J.; Mohammed, K.A.; Andjelic, C.D.; Barrows, L.R.; Ireland, C.M. Theopapuamide, a cyclic depsipeptide from a Papua New Guinea lithistid sponge Theonella swinhoei. J. Nat. Prod. 2006, 69, 1582–1586.
- Plaza, A.; Bifulco, G.; Keffer, J.L.; Lloyd, J.R.; Baker, H.L.; Bewley, C.A. Celebesides A–C and theopapuamides B–D, depsipeptides from an Indonesian sponge that inhibit HIV-1 entry. J. Org. Chem. 2009, 74, 504–512.
- Bewley, C.A.; He, H.; Williams, D.H.; Faulkner, D.J. Aciculitins A–C: Cytotoxic and antifungal cyclic peptides from the lithistid sponge Aciculites orientalis. J. Am. Chem. Soc. 1996, 118, 4314–4321.
- Ng-Choi, I.; Oliveras, À.; Feliu, L.; Planas, M. Solid-phase synthesis of biaryl cyclic peptides containing a histidine-phenylalanine linkage. Int. J. Pept. Res. Ther. 2020, 26, 695–707.
- Zampella, A.; D’Auria, M.V.; Paloma, L.G.; Casapullo, A.; Minale, L.; Debitus, C.; Henin, Y. Callipeltin A, an anti-HIV cyclic depsipeptide from the New Caledonian Lithistida sponge Callipelta sp. J. Am. Chem. Soc. 1996, 118, 6202–6209.
- Liang, B.; Carroll, P.J.; Joullié, M.M. Progress toward the total synthesis of callipeltin A (I): Asymmetric synthesis of (3S, 4 R)-3,4-dimethylglutamine. Org. Lett. 2000, 2, 4157–4160.
- Hansen, D.B.; Wan, X.; Carroll, P.J.; Joullié, M.M. Stereoselective synthesis of four stereoisomers of β-methoxytyrosine, a component of callipeltin A. J. Org. Chem. 2005, 70, 3120–3126.
- Kikuchi, M.; Konno, H. Total synthesis of callipeltin B and M, peptidyl marine natural products. Org. Lett. 2014, 16, 4324–4327.
- Zampella, A.; D’Auria, M.V. Stereoselective synthesis of (2R,3R,4R)-3-hydroxy-2,4,6-trimethylheptanoic acid and determination of the absolute stereochemistry of the natural product from callipeltin A. Tetrahedron Asymmetry 2002, 13, 1237–1239.
- Daletos, G.; Kalscheuer, R.; Koliwer-Brandl, H.; Hartmann, R.; De Voogd, N.J.; Wray, V.; Lin, W.; Proksch, P. Callyaerins from the marine sponge Callyspongia aerizusa: Cyclic peptides with antitubercular activity. J. Nat. Prod. 2015, 78, 1910–1925.
- Zhang, S.; De Leon Rodriguez, L.M.; Leung, I.K.; Cook, G.M.; Harris, P.W.; Brimble, M.A. Total Synthesis and Conformational Study of Callyaerin A: Anti-Tubercular Cyclic Peptide Bearing a Rare Rigidifying (Z)-2,3-Diaminoacrylamide Moiety. Angew. Chem. Int. Ed. 2018, 57, 3631–3635.
- Clark, D.P.; Carroll, J.; Naylor, S.; Crews, P. An antifungal cyclodepsipeptide, cyclolithistide A, from the sponge Theonella swinhoei. J. Org. Chem. 1998, 63, 8757–8764.
- Grieco, P.A.; Perez-Medrano, A. Total synthesis of the mixed peptide-polypropionate based cyclodepsipeptide (+)-geodiamolide B. Tetrahedron Lett. 1988, 29, 4225–4228.
- Chan, W.R.; Tinto, W.F.; Manchand, P.S.; Todaro, L.J. Stereostructures of geodiamolides A and B, novel cyclodepsipeptides from the marine sponge Geodia sp. J. Org. Chem. 1987, 52, 3091–3093.
- White, J.D.; Amedio, J.C., Jr. Total synthesis of geodiamolide A, a novel cyclodepsipeptide of marine origin. J. Org. Chem. 1989, 54, 736–738.
- Amagata, T.; Morinaka, B.I.; Amagata, A.; Tenney, K.; Valeriote, F.A.; Lobkovsky, E.; Clardy, J.; Crews, P. A chemical study of cyclic depsipeptides produced by a sponge-derived fungus. J. Nat. Prod. 2006, 69, 1560–1565.
- Yeo, S.-H.; Seo, H.-J.; Lim, D.-Y. Synthesis of halicylindramide a mimetics containing lactone isosteres. Bull. Korean Chem. Soc. 2011, 32, 2916–2920.
- Seo, H.; Lim, D. Total synthesis of Halicylindramide A. J. Org. Chem. 2009, 74, 906–909.
- Bellotta, F.; D’Auria, M.V.; Sepe, V.; Zampella, A. Synthetic studies on homophymine A: Stereoselective synthesis of (2R,3R,4R,6R)-3-hydroxy-2,4,6-trimethyloctanoic acid. Tetrahedron 2009, 65, 3659–3663.
- Ohtaka, J.; Hamajima, A.; Nemoto, T.; Hamada, Y. Efficient diastereoselective synthesis of (2R, 3R, 4R)-2-amino-3-hydroxy-4, 5-dimethylhexanoic acid, the lactone linkage unit of homophymine A. Chem. Pharm. Bull. 2013, 61, 245–250.
- Kobayashi, J.I.; Tsuda, M.; Nakamura, T.; Mikami, Y.; Shigemori, H. Hymenamides A and B, new proline-rich cyclic heptapeptides from the Okinawan marine sponge Hymeniacidon sp. Tetrahedron 1993, 49, 2391–2402.
- Shiki, Y.; Onai, M.; Sugiyama, D.; Osada, S.; Fujita, I.; Kodama, H. Synthesis and biological activities of cyclic peptide, hymenamide analogs. In Peptides for Youth; Springer: New York, NY, USA, 2009; pp. 323–324.
- Tsuda, M.; Shigemori, H.; Mikami, Y.; Kobayashi, J.i. Hymenamides C-E, new cyclic heptapeptides with two proline residues from the okinawan marine sponge hymeniacidon sp. Tetrahedron 1993, 49, 6785–6796.
- Chu, K.S.; Negrete, G.R.; Konopelski, J.P. Asymmetric total synthesis of (+)-jasplakinolide. J. Org. Chem. 1991, 56, 5196–5202.
- Grieco, P.A.; Hon, Y.S.; Perez-Medrano, A. Convergent, enantiospecific total synthesis of the novel cyclodepsipeptide (+)-jasplakinolide (jaspamide). J. Am. Chem. Soc. 1988, 110, 1630–1631.
- Ghosh, A.K.; Moon, D.K. Enantioselective total synthesis of (+)-jasplakinolide. Org. Lett. 2007, 9, 2425–2427.
- Davis, R.A.; Mangalindan, G.C.; Bojo, Z.P.; Antemano, R.R.; Rodriguez, N.O.; Concepcion, G.P.; Samson, S.C.; de Guzman, D.; Cruz, L.J.; Tasdemir, D. Microcionamides A and B, bioactive peptides from the Philippine sponge Clathria (Thalysias) abietina. J. Org. Chem. 2004, 69, 4170–4176.
- Bewley, C.A.; Debitus, C.; Faulkner, D.J. Microsclerodermins A and B. Antifungal cyclic peptides from the lithistid sponge Microscleroderma sp. J. Am. Chem. Soc. 1994, 116, 7631–7636.
- Schmidt, E.W.; Faulkner, D.J. Microsclerodermins C-E, antifungal cyclic peptides from the lithistid marine sponges Theonella sp. and Microscleroderma sp. Tetrahedron 1998, 54, 3043–3056.
- Qureshi, A.; Colin, P.L.; Faulkner, D.J. Microsclerodermins F–I, Antitumor and Antifungal Cyclic Peptides from the Lithistid Sponge Microscleroderma sp. Tetrahedron 2000, 56, 3679–3685.
- Zhang, X.; Jacob, M.R.; Rao, R.R.; Wang, Y.-H.; Agarwal, A.K.; Newman, D.J.; Khan, I.A.; Clark, A.M.; Li, X.-C. Antifungal cyclic peptides from the marine sponge Microscleroderma herdmani. Med. Chem. Res. 2012, 2, 7.
- Zhu, J.; Ma, D. Total synthesis of microsclerodermin E. Angew. Chem. Int. Ed. 2003, 42, 5348–5351.
- Santhakumar, G.; Payne, R.J. Studies toward the Total Synthesis and Stereochemical Assignment of Microspinosamide. Aust. J. Chem. 2016, 68, 1885–1889.
- Ramamoorthy, G.; Acevedo, C.M.; Alvira, E.; Lipton, M.A. Synthesis and spectroscopic correlation of the diastereoisomers of 2,3-dihydroxy-2,6,8-trimethyldeca-(4Z,6E)-dienoic acid: Implications for the structures of papuamides A–D and mirabamides A–D. Tetrahedron Asymmetry 2008, 19, 2546–2554.
- Okada, Y.; Matsunaga, S.; van Soest, R.W.; Fusetani, N. Nagahamide A, an antibacterial depsipeptide from the marine sponge Theonella swinhoei. Org. Lett. 2002, 4, 3039–3042.
- Mohapatra, D.K.; Chaudhuri, S.R.; Sahoo, G.; Gurjar, M.K. Stereoselective synthesis of the polyketide chain of nagahamide A. Tetrahedron Asymmetry 2006, 17, 2609–2616.
- D’Auria, M.V.; Paloma, L.G.; Minale, L.; Zampella, A.; Debitus, C.; Perez, J. Neosiphoniamolide A, a novel cyclodepsipeptide, with antifungal activity from the marine sponge Neosiphonia superstes. J. Nat. Prod. 1995, 58, 121–123.
- Makino, K.; Nagata, E.; Hamada, Y. Synthesis of tripeptide hydrolysate from papuamide A: Determination of absolute stereostructure of β-methoxytyrosine. Tetrahedron Lett. 2005, 46, 6827–6830.
- Okamoto, N.; Hara, O.; Makino, K.; Hamada, Y. Diastereoselective synthesis of all stereoisomers of β-methoxytyrosine, a component of papuamides. J. Org. Chem. 2002, 67, 9210–9215.
- Xie, W.; Ding, D.; Zi, W.; Li, G.; Ma, D. Total synthesis and structure assignment of papuamide B, a potent marine cyclodepsipeptide with anti-HIV properties. Angew. Chem. 2008, 120, 2886–2890.
- Makino, K.; Nagata, E.; Hamada, Y. Practical synthesis of (2S,3R)-3hydroxy-3-methylproline, a constituent of papuamides, using a diastereoselective tandem Michael-aldol reaction. Tetrahedron Lett. 2005, 46, 8159–8162.
- Gulavita, N.K.; Gunasekera, S.P.; Pomponi, S.A.; Robinson, E.V. Polydiscamide A: A new bioactive depsipeptide from the marine sponge Discodermia sp. J. Org. Chem. 1992, 57, 1767–1772.
- Santhakumar, G.; Payne, R.J. Total synthesis of polydiscamides B, C, and D via a convergent native chemical ligation–oxidation strategy. Org. Lett. 2014, 16, 4500–4503.
- Shin, H.J.; Rashid, M.A.; Cartner, L.K.; Bokesch, H.R.; Wilson, J.A.; McMahon, J.B.; Gustafson, K.R. Stellettapeptins A and B, HIV-inhibitory cyclic depsipeptides from the marine sponge Stelletta sp. Tetrahedron Lett. 2015, 56, 4215–4219.
- Dahiya, R.; Singh, S.; Sharma, A.; Chennupati, S.V.; Maharaj, S. First total synthesis and biological screening of a proline-rich cyclopeptide from a Caribbean marine sponge. Mar. Drugs 2016, 14, 228.
- Bewley, C.A.; Faulkner, D.J. Theonegramide, an antifungal glycopeptide from the Philippine lithistid sponge Theonella swinhoei. J. Org. Chem. 1994, 59, 4849–4852.
- Tohdo, K.; Hamada, Y.; Shioiri, T. Theonellamide F synthetic studies. Stereoselective synthesis of (3S, 4S, 5E, 7E)-3-amino-8-(4-bromophenyl)-4-hydroxy-6-methyl-5, 7-octadienoic acid (aboa). Tetrahedron Lett. 1992, 33, 2031–2034.
- Tsuda, M.; Shimbo, K.; Kubota, T.; Mikami, Y.; Kobayashi, J.i. Two theonellapeptolide congeners from marine sponge Theonella sp. Tetrahedron 1999, 55, 10305–10314.
- Kuranaga, T.; Enomoto, A.; Tan, H.; Fujita, K.; Wakimoto, T. Total synthesis of theonellapeptolide Id. Org. Lett. 2017, 19, 1366–1369.
- Stincone, P.; Brandelli, A. Marine bacteria as source of antimicrobial compounds. Crit. Rev. Biotechnol. 2020, 40, 306–319.
More
Information
Subjects:
Infectious Diseases
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.0K
Entry Collection:
Peptides for Health Benefits
Revisions:
2 times
(View History)
Update Date:
04 Jul 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





