
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Navid Sobhani | + 1447 word(s) | 1447 | 2021-02-25 07:36:05 | | | |
| 2 | Camila Xu | Meta information modification | 1447 | 2021-03-18 03:48:04 | | |
Video Upload Options
CD44 is a receptor described as a single span transmembrane glycoprotein without kinase activity whose ubiquitous and constitutive expression has been observed on many different cells.
1. ‘Facts Check’—CD44 Receptor
CD44 can exist as an integral component of the extracellular matrix (ECM) or as a soluble protein found in body fluids [1]. Each of these protein forms plays biological roles, which may be shared or distinct [1].
The membrane form of CD44 is a receptor that binds ECM ligands, such as hyaluronan (HA), collagen, osteopontin, laminin, fibronectin, and others. The structure of CD44 in its standard (CD44s) and variant (CD44v) form is presented in Figure 1. CD44 is encoded by a single gene, which in humans is composed of 20 exons [2]. The schematic CD44 gene structure is presented in Figure 2. CD44s is composed of the homologous N-terminal and C-terminal domains only. The N-terminal domain is encoded by exons 1–5, whereas the C-terminal domain is encoded by exons 16–20. The molecular mass of CD44s is usually between 80–85 kDa. Multiple variant forms (CD44v, v1–v10) are generated by alternative splicing of exons 6–15, and are composed of both the N- and C-terminal domains and an additional variant domain(s). While CD44s is found in a wide variety of tissues, the CD44v isoform seems to have a much-restricted distribution and is observed to be present in keratinocytes, activated lymphocytes, macrophages, and some epithelial cells [3]. In both CD44 forms, the N-terminal domain binds the physiological ligands. CD44v isoforms are enriched in binding motifs, which promote the interactions with microenvironment components and may serve as co-receptors for growth factors, such as EGF and HGF [4]. Structural diversity of CD44 is also generated by posttranslational modification, such as phosphorylation, glycosylation, and attachment of glycosaminoglycans [1][5]. Resting cells show a low-affinity for binding CD44 ligands; however, cellular activation can induce a transition of CD44 to a high-affinity state that mediates binding to HA. The transition from the “inactive” low-affinity state to the “active” high-affinity state of CD44 can be induced by soluble factors, including cytokines [6][7]. Activation of CD44 leads to clustering and initiation of intracellular signaling pathways, such as ERK1, 2, AKT, FAK, Rho, Src, etc. (reviewed in [8]). Tumor cells express CD44 in a high-affinity state with the capacity for constitutive binding of HA [9]. The soluble and/or integral with ECM forms of CD44 are generated by metalloproteinase-inflicted (mainly MT1- and MT3-MMPs) shedding from a cell’s membrane [10]. The soluble CD44 protein can also be also produced by alternative splicing [11]. This form has been detected in the circulation and other body fluids, such as lymphatic, arthritic synovial, and bronchoalveolar fluids, as well as in cancer patients’ serum [12][13][14]. Soluble CD44 overexpression blocks cancer cell adhesion to HA [15] and is associated with aggressive growth and unfavorable prognosis for a patient.
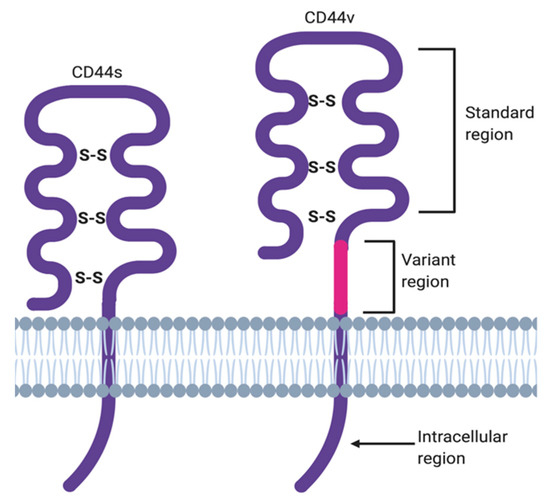
Figure 1. CD44 structure in its standard (CD44s) and variant (CD44v) forms.
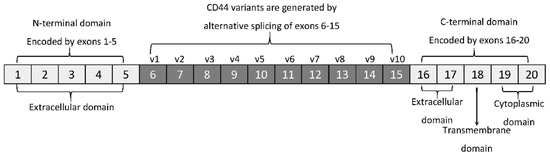
Figure 2. CD44 gene structure. CD44s form is encoded by exons 1–5 and 16–20 only, whereas CD44v form undergoes alternative splicing (exons 6–15).
2. Hyaluronan—The Major Ligand of CD44
As mentioned above, CD44 binds to a number of ligands; however, hyaluronan (hyaluronic acid, HA) is its most specific ligand that is recognizable by both standard and variant forms of this receptor [8]. HA is an anionic, non-sulfated polysaccharide that belongs to glycosaminoglycans. Its basic disaccharide monomer is a D-glucuronic acid linked with N-acetyl-D-glucosamine by glycosidic bonds (β1–4, β1–3). The schematic structure of HA is presented in Figure 3. The molecular weight of HA varies from 103 to 107 Da and depends on its tissue origin. In contrast to other glycosaminoglycans, HA is synthesized by three HA synthases (HAS 1–3) at the cell surface (not in Golgi apparatus) [16] and is released into the extracellular space. HA is a major component of ECM, which supports tissue organization and contributes to crucial processes, such as cell migration, proliferation, survival, etc. [17].
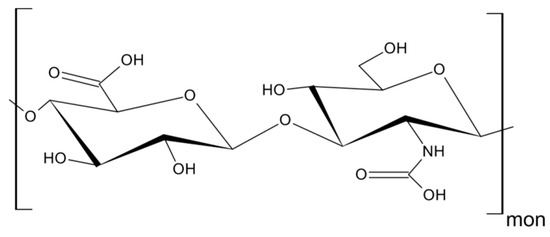
Figure 3. Structure of hyaluronan (HA).
The downstream effects of HA binding depend on its molecular weight. Under normal circumstances, HA, in its “native state”, represents a high-molecular weight protein of about 1000 kDa [18] with anti-angiogenic, anti-inflammatory, and proliferation inhibiting activities [19]. On the other hand, HA in its ‘fragmented state’, with a molecular weight below 500 kDa, stimulates proliferation and migration of cells, induces an immune response, e.g., by increasing expression of inflammatory genes in human macrophages [18][20], the formation of immunosuppressive macrophages [21], death of activated T cells [22], and augments angiogenesis [19].
A number of studies have shown that HA is produced in abundance by malignant and stromal cells supporting the growth of tumors (breast [23], bladder [24], prostate [25] colorectal cancer [26]), and correlates with tumor aggressiveness in humans [21][26]. HA facilitates tumor cell proliferation, cell-to-cell adhesion and protects them from the immune cell response [27]. The balance between HA contradictory activities is governed by a complex system of HA synthases and hyaluronidases (Hyal), which regulate its degradation. Tumor cells have been reported to secrete excess amounts of hyaluronidases with the ability to cleave high molecular weight HA into smaller fragments (10–40 oligomers) [28], which, in turn, promoted CD44 cleavage [29]. Enhanced cleavage of CD44 was shown to facilitate tumor cell motility, which has been observed in many human tumors, including breast, colon, gastric, ovarian, lung, and others [28]. It has been shown that HA is involved in cancer cell, lymphocyte, and neutrophil motility in a CD44-dependent manner, however, the expression of this receptor alone was insufficient for chemotaxis towards HA [30][31]. Data on the binding efficiency of HA to CD44s and CD44v are conflicting. Bennett et al. reported that CD44s binds HA more efficiently than CD44v [32], however, other authors presented convincing data showing HA’s affinity to both receptors to be similar [33].
3. CD44-HA Interactions
There is quite an extensive number of surface receptors that recognize either the soluble or molecular form of HA. These include proteins, such as CD44, RHAMM, TSG-6/TNFIP6, Brevican, SHAP, LYVE1, TLRs, and others [17]. The RHAMM and TLR2,4 receptors preferentially bind smaller HA molecules (below 7 kDa), whereas CD44 requires at least six monosaccharides for the process to occur [reviewed in [34]. CD44 contains at least three HA binding sites on the hair loop (encoded by exon 2 and 5). CD44 molecules have been reported to be present in three functional stages: Constitutive binding of HA (most of the tumor cells), non-binding or non-binding until activated by physiological stimuli [8][35]. The affinity of HA to CD44 is cell-type dependent, regulated by glycosylation in the receptor’s extracellular domain and the phosphorylation of the serine residue in the cytoplasmic tail [36]. All forms of CD44 (standard or variants) can bind HA. In the case of immune cells, CD44 was described as the only receptor that has been demonstrated to bind HA [31].
Upon HA binding to the intercellular domain, CD44 undergoes conformational changes resulting in the recruitment of adaptor proteins (ERM, Src, etc.) to its intracellular domain, which, in turn, leads to the activation of a number of signaling pathways involved in cell proliferation, adhesion, migration, invasion and metabolic shift [37][38]. Table 1 summarizes all the currently known signal transduction mechanisms and their outcomes associated with CD44 molecule in cancer cells. Moreover, CD44v isoforms can also serve as co-receptors with the ability to sequester growth factors (i.e., HGF, FGF, EGF) at the cell surface and presenting them to tyrosine kinase receptor (RTK) complexes (i.e., c-Met, EGFR), which in turn, upon activation, can initiate an appropriate signaling pathway. Figure 4 depicts possible intercellular signaling mediated by the CD44 receptor in cancer cells.
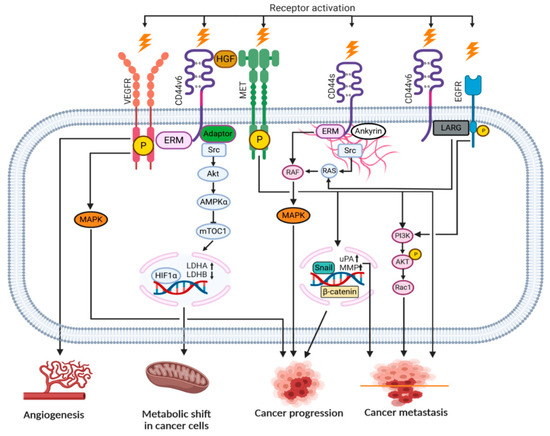
Figure 4. Signaling pathways engaging the CD44 receptor in cancer cells (according to [8] with own modifications).
Table 1. Association of the CD44 molecule with signaling pathways in cancer cells.
| Signaling Pathway | Outcome | Reference |
|---|---|---|
| CD44s | ERM/MAPK | Cell division, proliferation |
| PI3K/AKT/NFκB | Inflammatory cytokine release | |
| Migration of ICD fragment into the nucleus | Metastasis, survival | |
| RHAMM/cSrc/RAS | Migration | |
| CD44v | ERM/FAK/Paxillin | Angiogenesis |
| Src/AKT/HIF1α | Metabolic shift (glycolysis increase) | |
| Met/PI3K/AKT | Cell invasion | |
| Met/Snail/MMP | Cell division, proliferation |
CD44–HA interactions were defined as important for many types of cells. For example, macrophages exhibited preferential migration towards HA-enriched stromal structures in breast cancer [38]. It was also shown that small HA induced immunophenotypic maturation of human monocyte-derived dendritic cells and an increase in their pro-inflammatory cytokine (IL-1, TNF, and IL-12) production [39]. An elevated expression of CD44 has been reported on activated T cells [18]. The CD44-dependent adhesion mechanism is likely to be the most important for mobilizing the effector T cells at sites of infection and inflammation, because the expression of CD44 is upregulated whereas L-selectin (CD62L), the lymphocyte-expressed selectin, is downregulated in these cases [40]. Activation of CD44 influenced tumor cell metabolism by inducing Hypoxia-inducible factor 1α (HIF1α) binding to nuclear DNA to increase glycolysis, in turn, rendering a metabolic shift in cancer cells [41].
References
- Cichy, J.; Puré, E. The liberation of CD44. J. Cell Biol. 2003, 161, 839–843.
- Yan, Y.; Zuo, X.; Wei, D. Concise review: Emerging role of CD44 in cancer stem cells: A promising biomarker and therapeutic target. Stem Cells Transl. Med. 2015, 4.
- Sneath, R.J.S.; Mangham, D.C. The normal structure and function of CD44 and its role in neoplasia. J. Clin. Pathol. Mol. Pathol. 1998, 51, 191.
- Van Der Voort, R.; Taher, T.E.I.; Wielenga, V.J.M.; Spaargaren, M.; Prevo, R.; Smit, L.; David, G.; Hartmann, G.; Gherardi, E.; Pals, S.T. Heparan sulfate-modified CD44 promotes hepatocyte growth factor/scatter factor-induced signal transduction through the receptor tyro sine kinase c-Met. J. Biol. Chem. 1999.
- Gao, T.; Wen, T.; Ge, Y.; Liu, J.; Yang, L.; Jiang, Y.; Dong, X.; Liu, H.; Yao, J.; An, G. Disruption of Core 1-mediated O-glycosylation oppositely regulates CD44 expression in human colon cancer cells and tumor-derived exosomes. Biochem. Biophys. Res. Commun. 2020.
- Cichy, J.; Puré, E. Cytokines regulate the affinity of soluble CD44 for hyaluronan. FEBS Lett. 2004.
- Cichy, J.; Puré, E. Oncostatin M and transforming growth factor-β1 induce post-translational modification and hyaluronan binding to CD44 in lung-derived epithelial tumor cells. J. Biol. Chem. 2000.
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64.
- Naor, D.; Sionov, R.V.; Ish-Shalom, D. CD44: Structure, function, and association with the malignant process. Adv. Cancer Res. 1997, 71, 241–319.
- Kajita, M.; Itoh, Y.; Chiba, T.; Mori, H.; Okada, A.; Kinoh, H.; Seiki, M. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 2001.
- Yu, Q.; Toole, B.P. A new alternatively spliced exon between v9 and v10 provides a molecular basis for synthesis of soluble CD44. J. Biol. Chem. 1996.
- Katoh, S.; McCarthy, J.B.; Kincade, P.W. Characterization of soluble CD44 in the circulation of mice: Levels are affected by immune activity and tumor growth. J. Immunol. 1994, 153.
- Katoh, S.; Taniguchi, H.; Matsubara, Y.; Matsumoto, N.; Fukushima, K.; Kadota, J.; Matsukura, S.; Kohno, S. Overexpression of CD44 on alveolar eosinophils with high concentrations of soluble CD44 in bronchoalveolar lavage fluid in patients with eosinophilic pneumonia. Allergy Eur. J. Allergy Clin. Immunol. 1999, 54.
- Shi, M.; Dennis, K.; Peschon, J.J.; Chandrasekaran, R.; Mikecz, K. Antibody-induced shedding of CD44 from adherent cells is linked to the assembly of the cytoskeleton. J. Immunol. 2001, 167.
- Päll, T.; Pink, A.; Kasak, L.; Turkina, M.; Anderson, W.; Valkna, A.; Kogerman, P. Soluble CD44 interacts with intermediate filament protein vimentin on endothelial cell surface. PLoS ONE 2011, 6, 1–11.
- Schwertfeger, K.L.; Cowman, M.K.; Telmer, P.G.; Turley, E.A.; McCarthy, J.B. Hyaluronan, inflammation, and breast cancer progression. Front. Immunol. 2015, 6, 236.
- Toole, B.P. Hyaluronan-CD44 interactions in cancer: Paradoxes and possibilities. Clin. Cancer Res. 2009, 15, 7462–7468.
- Jiang, D.; Liang, J.; Noble, P.W. Hyaluronan as an immune regulator in human diseases. Physiol. Rev. 2011, 91, 221–264.
- Tolg, C.; McCarthy, J.B.; Yazdani, A.; Turley, E.A. Hyaluronan and RHAMM in Wound Repair and the “cancerization” of Stromal Tissues. BioMed Res. Int. 2014, 2014.
- McKee, C.M.; Penno, M.B.; Cowman, M.; Burdick, M.D.; Strieter, R.M.; Bao, C.; Noble, P.W. Hyaluronan (HA) fragments induce chemokine gene expression in alveolar macrophages: The role of HA size and CD44. J. Clin. Invest. 1996.
- Kuang, D.M.; Wu, Y.; Chen, N.; Cheng, J.; Zhuang, S.M.; Zheng, L. Tumor-derived hyaluronan induces formation of immunosuppressive macrophages through transient early activation of monocytes. Blood 2007.
- Ruffell, B.; Johnson, P. Hyaluronan induces cell death in activated T cells through CD44. J. Immunol. 2008.
- Auvinen, P.; Rilla, K.; Tumelius, R.; Tammi, M.; Sironen, R.; Soini, Y.; Kosma, V.M.; Mannermaa, A.; Viikari, J.; Tammi, R. Hyaluronan synthases (HAS1-3) in stromal and malignant cells correlate with breast cancer grade and predict patient survival. Breast Cancer Res. Treat. 2014.
- Lokeshwar, V.B.; Öbek, C.; Soloway, M.S.; Block, N.L. Tumor-associated hyaluronic acid: A new sensitive and specific urine marker for bladder cancer. Cancer Res. 1997, 57, 773–777.
- Josefsson, A.; Adamo, H.; Hammarsten, P.; Granfors, T.; Stattin, P.; Egevad, L.; Laurent, A.E.; Wikström, P.; Bergh, A. Prostate cancer increases hyaluronan in surrounding nonmalignant stroma, and this response is associated with tumor growth and an unfavorable outcome. Am. J. Pathol. 2011.
- Ropponen, K.; Tammi, M.; Parkkinen, J.; Eskelinen, M.; Tammi, R.; Lipponen, P.; Ågren, U.; Alhava, E.; Kosma, V.M. Tumor cell-associated hyaluronan as an unfavorable prognostic factor in colorectal cancer. Cancer Res. 1998, 58, 342–347.
- McBride, W.H.; Bard, J.B.L. Hyaluronidase-sensitive halos around adherent cells: Their role in blocking lymphocyte-mediated cytolysis. J. Exp. Med. 1979.
- Sugahara, K.N.; Hirata, T.; Hayasaka, H.; Stern, R.; Murai, T.; Miyasaka, M. Tumor cells enhance their own CD44 cleavage and motility by generating hyaluronan fragments. J. Biol. Chem. 2006.
- Sugahara, K.N.; Murai, T.; Nishinakamura, H.; Kawashima, H.; Saya, H.; Miyasaka, M. Hyaluronan oligosaccharides induce CD44 cleavage and promote cell migration in CD44-expressing tumor cells. J. Biol. Chem. 2003.
- Tzircotis, G.; Thorne, R.F.; Isacke, C.M. Chemotaxis towards hyaluronan is dependent on CD44 expression and modulated by cell type variation in CD44-hyaluronan binding. J. Cell Sci. 2005.
- Lee-Sayer, S.S.M.; Dong, Y.; Arif, A.A.; Olsson, M.; Brown, K.L.; Johnson, P. The where, when, how and why of hyaluronan binding by immune cells. Front. Immunol. 2015, 6, 150.
- Bennett, K.L.; Modrell, B.; Greenfield, B.; Bartolazzi, A.; Stamenkovic, I.; Peach, R.; Jackson, D.G.; Spring, F.; Aruffo, A. Regulation of CD44 binding to hyaluronan by glycosylation of variably spliced exons. J. Cell Biol. 1995.
- Raman, P.S.; Alves, C.S.; Wirtz, D.; Konstantopoulos, K. Distinct kinetic and molecular requirements govern CD44 binding to hyaluronan versus fibrin(ogen). Biophys. J. 2012.
- Campo, G.M.; Avenoso, A.; Micali, A.; Nastasi, G.; Squadrito, F.; Altavilla, D.; Bitto, A.; Polito, F.; Rinaldi, M.G.; Calatroni, A.; et al. High-molecular weight hyaluronan reduced renal PKC activation in genetically diabetic mice. Biochim. Biophys. Acta Mol. Basis Dis. 2010, 1802.
- Lesley, J.; Hascall, V.C.; Tammi, M.; Hyman, R. Hyaluronan binding by cell surface CD44. J. Biol. Chem. 2000.
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Front. Immunol. 2015, 6, 201.
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45.
- Kobayashi, N.; Miyoshi, S.; Mikami, T.; Koyama, H.; Kitazawa, M.; Takeoka, M.; Sano, K.; Amano, J.; Isogai, Z.; Niida, S.; et al. Hyaluronan deficiency in tumor stroma impairs macrophage trafficking and tumor neovascularization. Cancer Res. 2010, 70.
- Termeer, C.; Sleeman, J.P.; Simon, J.C. Hyaluronan—Magic glue for the regulation of the immune response? Trends Immunol. 2003, 24, 112–114.
- Baaten, B.J.G.; Li, C.R.; Bradley, L.M. Multifaceted regulation of T cells by CD44. Commun. Integr. Biol. 2010, 3.
- Nam, K.; Oh, S.; Shin, I. Ablation of CD44 induces glycolysis-to-oxidative phosphorylation transition via modulation of the c-Src-Akt-LKB1-AMPK pathway. Biochem. J. 2016, 473, 3013–3030.




