
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Shahid Iqbal | + 1522 word(s) | 1522 | 2020-09-25 11:39:03 | | | |
| 2 | Rita Xu | -750 word(s) | 772 | 2021-02-18 03:21:30 | | |
Video Upload Options
The problem of pistil abortion in Japanese apricot is very common but has not been well studied. Therefore, in the current study, we used RNA-Seq to investigate the molecular mechanism underlying pistil abortion. This entry provides a theoretical basis for stimulating flower and pistil development and has practical significance for improving yield and quality of Japanese apricot.
1. Introduction
Japanese apricot (Prunus mume Sieb. et Zucc) is an important fruit and ornamental plant. It has great economic importance, high profit and world market demand, and it is used in value-added products like jams, pickles, etc. [1][2]. Flower development is a fundamental stage in the life cycle of the plant and plays a significant role in the sexual reproduction process [3]. Pistil abortion is a widespread phenomenon occurring in fruit plants and has been discussed in different fruit crops like pomegranate, Xanthoceras sorbifolia and olive [4][5][6]. In Japanese apricot, the problem of pistil abortion is very common and causes serious losses, such as decrease in fruit quality and yield, thus limiting the apricot growing industry. The abortive pistils are characterized as withered or absent (no pistils) [7]. Several proteomic studies have been performed and found that glucose, starch and photosynthesis metabolisms were associated with pistil abortion [8].
In fruit trees, pistil abortion may result from improper ovary development, style formation and fertilization during flower development [9]. The rate of pistil abortion depends on the type of apricot cultivar, as the abortion rate in Japanese apricot fruit can reach up to 76.3% [10], while in some cultivars of Xinjiang apricot the pistil abortion rate can range from 60.4% to 99.64% [11]. Numerous studies showed that pistil abortion might be controlled through many factors such as the environment (light, humidity, temperature), endogenous substances and signals associated with various biological and cellular processes during floral organ formation [12].
Plant hormones are actively involved in floral transition and floral organ development [13], and they play a regulatory role in physiological processes of flower bud development including floral bud initiation [14], differentiation [15] and dormancy [16]. As an excellent growth regulator, abscisic acid (ABA) is an active hormone involved in floral time transition [17]. In Arabidopsis, cytokinin (CK) promotes flowering [18][19], while gibberellin (GA) promotes cell division and growth [20]. Cytokinin affects the expression of PIN1 protein through promoting the expression of AG and SPL genes, thereby affecting normal pistil and ovule development [21]. Therefore, the above related information confirms the substantial involvement of plant hormones during pistil abortion.
RNA-Seq technology is extensively used to determine gene expression and the associated genetic network during flower development [22], and it has previously been used in studies of sugar apple [23], pomegranate [4] and olive [24]. The problem of pistil abortion in Japanese apricot is very common but has not been well studied. Therefore, in the current study, we used RNA-Seq to investigate the molecular mechanism underlying pistil abortion.
2. An Overview of RNA-Seq Libraries
In the current study, six different cDNA libraries were constructed for normal and abortive pistils and sequenced using Illumina sequencing, producing an average of 65.8 M raw reads per sample. After the removal of raw reads and adopter sequences, 63.27 M clean reads per sample were obtained and mapped to the P. mume genome. After mapping to the reference genome, a total of 86.64% mapped reads were obtained from each sample, while uniquely mapped reads were 63.94% per sample (Table 1).
Table 1. An overview of sequencing assembly in normal and abortive pistils of Japanese apricot.
| Sample Name | Total Raw Reads (Mb) | Total Clean Reads (Mb) | Clean Reads Q20 (%) | Clean Reads Q30 (%) | Clean Reads Ratio (%) | Total Mapping (%) | Uniquely Mapping (%) | Accession Number |
|---|---|---|---|---|---|---|---|---|
| NP-1 | 62.47 | 61.66 | 97.21 | 92.63 | 98.71 | 86.19 | 61.90 | SRR12234382 |
| NP-2 | 62.52 | 61.72 | 97.22 | 92.67 | 98.80 | 84.95 | 61.07 | SRR12234384 |
| NP-3 | 62.47 | 61.74 | 97.12 | 92.41 | 98.83 | 88.24 | 62.97 | SRR12234380 |
| AP-1 | 69.96 | 65.16 | 97.48 | 89.81 | 93.13 | 86.42 | 65.53 | SRR12234385 |
| AP-2 | 67.47 | 63.54 | 97.59 | 90.16 | 94.18 | 87.77 | 66.69 | SRR12234383 |
| AP-3 | 69.96 | 65.83 | 97.61 | 90.22 | 94.08 | 86.28 | 65.53 | SRR12234381 |
3. DEG Analysis of Normal and Abortive Pistils
Hierarchical clustering was performed for differentially expressed genes on the basis of their fold change values (log2fc), and the clustering is shown in Figure 1A. Pairwise comparison was used to analyze the regulation (up/down) and gene expression profiles between AP and NP. A total of 3882 unigenes (2033 up-regulated and 1849 down-regulated) were identified from normal and abortive pistil comparisons (Figure 1B). The volcano plot shows the expression level and overall distribution of differentially expressed genes between these two groups (Figure 1C).
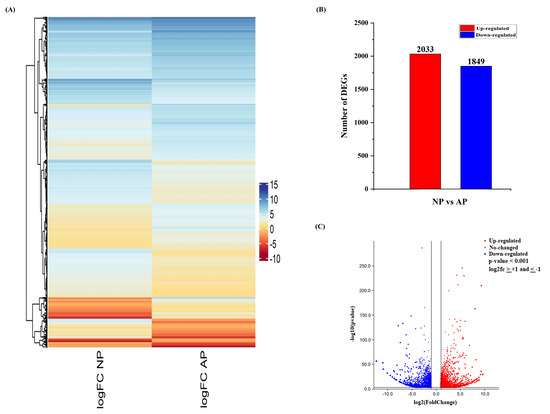
Figure 1. An overview of differentially expressed genes (DEGs) involved in normal and abortive pistils of Japanese apricot. (A) Heatmap showing the differential expression pattern of the DEGs. The color scale shows the gene expression values (log2fc). (B) Up- and down-regulation of differentially expressed genes. (C) Volcano plot showing the differentially expressed genes. The x-axis represents the log2 fold change conversion of the values, and the y-axis represents the significance value after –log10 conversion. Red shows up-regulated DEGs, blue shows down-regulated DEGs, while grey represents no DEGs.
References
- Adachi, M.; Suzuki, Y.; Mizuta, T.; Osawa, T.; Adachi, T.; Osaka, K.; Suzuki, K.; Shiojima, K.; Arai, Y.; Masuda, K.; et al. The “Prunus mume Sieb. et Zucc” (Ume) is a Rich Natural Source of Novel Anti-Cancer Substance. J. Food Prop. 2007, 10, 375–384, doi:10.1080/10942910600547624.
- Chu, M. Chinese fruit tree: Prunus mume; China Forestry Publishing House: Beijing, China, 1999.
- Huang, Y.; Liu, L.; Huang, J.; Wang, Z.; Chen, F.-F.; Zhang, Q.; Zheng, B.; Chen, M. Use of transcriptome sequencing to understand the pistillate flowering in hickory (Carya cathayensis Sarg.). BMC Genom. 2013, 14, 691, doi:10.1186/1471-2164-14-691.
- Chen, L.; Zhang, J.; Li, H.; Niu, J.; Xue, H.; Liu, B.; Wang, Q.; Luo, X.; Zhang, F.; Zhao, D.; et al. Transcriptomic Analysis Reveals Candidate Genes for Female Sterility in Pomegranate Flowers. Plant Sci. 2017, 8, 8, doi:10.3389/fpls.2017.01430.
- Gao, S.M.; Ma, K.; Du, X.H.; Li, F.L. Advances in research on Xanthoceras sorbifolia. Bull Bot 2002, 19, 296–301.
- Reale, L.; Sgromo, C.; Ederli, L.; Pasqualini, S.; Orlandi, F.; Fornaciari, M.; Ferranti, F.; Romano, B. Morphological and cytological development and starch accumulation in hermaphrodite and staminate flowers of olive (Olea europaea L.). Plant Reprod. 2009, 22, 109–119, doi:10.1007/s00497-009-0096-1.
- Hou, J.-H.; Gao, Z.-H.; Zhang, Z.; Chen, S.-M.; Ando, T.; Zhang, J.-Y.; Wang, X. Isolation and Characterization of an AGAMOUS Homologue PmAG from the Japanese Apricot (Prunus mume Sieb. et Zucc.). Plant Mol. Boil. Rep. 2010, 29, 473–480, doi:10.1007/s11105-010-0248-3.
- Wang, S. Preliminary Studies on Differences of Related Characteristics between Perfect Flower and Imperfect Flower and Protemics in Japanese Apricot; Nanjing Agricultural University: Nanjing, China, 2008.
- Wetzstein, H.Y.; Ravid, N.; Wilkins, E.; Martinelli, A.P. A Morphological and Histological Characterization of Bisexual and Male Flower Types in Pomegranate. Am. Soc. Hortic. Sci. 2011, 136, 83–92, doi:10.21273/jashs.136.2.83.
- Shi, T.; Zhang, Q.; Gao, Z.; Zhang, Z.; Zhuang, W. Analyses on pistil differentiation process and related biochemical indexes of two cultivars of Prunus mume. Plant Resour. Environ. 2011, 20, 35–41.
- Bai, Z.; Feng, J.; Li, W.; Hu, Y.; Cao, X.; Sun, J. Investigation of pistil abortion rate of five apricot cultivars in Xinjiang. Xinjiang Agric. Sci. 2012, 49, 1805–1809.
- Dokoozlian, N.K. Grape berry growth and development. Raisin Prod. Man. 2000, 3393, 30.
- Davis, S.J. Integrating hormones into the floral-transition pathway ofArabidopsis thaliana. Plant Cell Environ. 2009, 32, 1201–1210, doi:10.1111/j.1365-3040.2009.01968.x.
- Zhang, D.; Ren, L.; Yue, J.-H.; Wang, L.; Zhuo, L.-H.; Shen, X. A comprehensive analysis of flowering transition in Agapanthus praecox ssp. orientalis (Leighton) Leighton by using transcriptomic and proteomic techniques. Proteom. 2013, 80, 1–25, doi:10.1016/j.jprot.2012.12.028.
- Fu, C.; Huang, N.; Li, S.; Zhao, Z.; Huang, Z.; Shi, Y.; Tang, F. Endogenous hormones contents and growth development in South Feng-shui pear. Southwest China J. Agric. Sci. 2014, 27, 276–279.
- Zhu, Y.; Li, Y.; Xin, D.; Chen, W.; Shao, X.; Wang, Y.; Guo, W. RNA-Seq-based transcriptome analysis of dormant flower buds of Chinese cherry (Prunus pseudocerasus). Gene 2015, 555, 362–376, doi:10.1016/j.gene.2014.11.032.
- Jacobsen, J.V.; Pearce, D.W.; Poole, A.T.; Pharis, R.P.; Mander, L.N. Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Plant 2002, 115, 428–441, doi:10.1034/j.1399-3054.2002.1150313.x.
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin Regulates the Activity of Reproductive Meristems, Flower Organ Size, Ovule Formation, and Thus Seed Yield in Arabidopsis thaliana. Plant Cell 2011, 23, 69–80, doi:10.1105/tpc.110.079079.
- Corbesier, L.; Prinsen, E.; Jacqmard, A.; Lejeune, P.; Van Onckelen, H.; Perilleux, C.; Bernier, G. Cytokinin levels in leaves, leaf exudate and shoot apical meristem of Arabidopsis thaliana during floral transition. Exp. Bot. 2003, 54, 2511–2517, doi:10.1093/jxb/erg276.
- Debeaujon, I. Gibberellin Requirement for Arabidopsis Seed Germination Is Determined Both by Testa Characteristics and Embryonic Abscisic Acid. Plant Physiol. 2000, 122, 415–424, doi:10.1104/pp.122.2.415.
- Bencivenga, S.; Simonini, S.; Benková, E.; Colombo, L. The Transcription Factors BEL1 and SPL Are Required for Cytokinin and Auxin Signaling During Ovule Development in Arabidopsis[W]. Plant Cell 2012, 24, 2886–2897, doi:10.1105/tpc.112.100164.
- Zhang, L.; Wang, L.; Yang, Y.; Cui, J.; Chang, F.; Wang, Y.; Zhang, X. Analysis of Arabidopsis floral transcriptome: Detection of new florally expressed genes and expansion of Brassicaceae-specific gene families. Plant Sci. 2015, 5, 802, doi:10.3389/fpls.2014.00802.
- Liu, K.; Feng, S.; Pan, Y.; Zhong, J.; Chen, Y.; Yuan, C.; Li, H. Transcriptome Analysis and Identification of Genes Associated with Floral Transition and Flower Development in Sugar Apple (Annona squamosa L.). Plant Sci. 2016, 7, doi:10.3389/fpls.2016.01695.
- Alagna, F.; Cirilli, M.; Galla, G.; Carbone, F.; Daddiego, L.; Facella, P.; Lopez, L.; Colao, C.; Mariotti, R.; Cultrera, N.G.M.; et al. Transcript Analysis and Regulative Events during Flower Development in Olive (Olea europaea L.). PLoS ONE 2016, 11, e0152943, doi:10.1371/journal.pone.0152943.




