
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | shijia wu | -- | 1639 | 2024-02-21 12:53:26 | | | |
| 2 | shijia wu | + 3134 word(s) | 4773 | 2024-02-25 02:52:19 | | | | |
| 3 | Lindsay Dong | -353 word(s) | 4420 | 2024-02-27 06:30:12 | | | | |
| 4 | Lindsay Dong | Meta information modification | 4420 | 2024-02-27 06:32:06 | | | | |
| 5 | Lindsay Dong | Meta information modification | 4420 | 2024-02-27 06:33:39 | | | | |
| 6 | Lindsay Dong | Meta information modification | 4420 | 2024-02-29 07:35:52 | | | | |
| 7 | Lindsay Dong | Meta information modification | 4420 | 2024-03-08 06:21:44 | | |
Video Upload Options
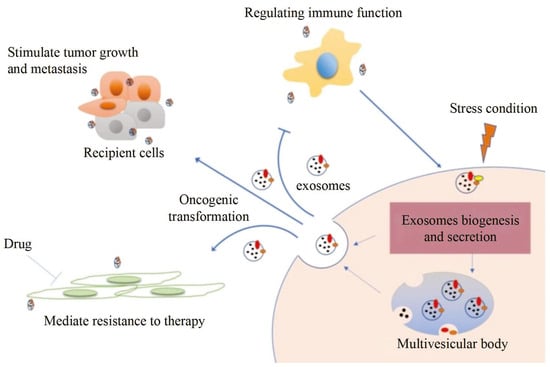
Exosomes distributed by extracellular vesicles carry various information highly consistent with cells, becoming a new type of biomarker for tumor screening. However, although conventional characterization technologies can quantify size and morphology for exosomes, they are limited in related fields such as function tracing, protein quantification at unit point, and microstructural information.
1. Introduction
2. Conventional Characterization Technologies
2.1. Tunable Resistive Pulse Sensing
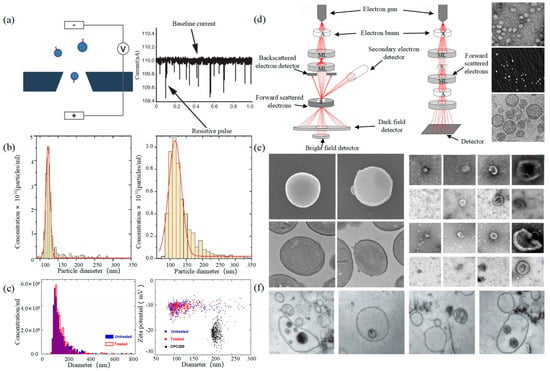
2.2. Electron Microscope
Electron Microscope (EM) is the most direct method to measure the size and morphology of a single EV [42]. It is divided into scanning electron microscope (SEM), transmission electron microscope (TEM), and cryo-transmission electron microscope (cryo-TEM). It is noted that EM can characterize the particle morphology and size of a single EV [53][54].
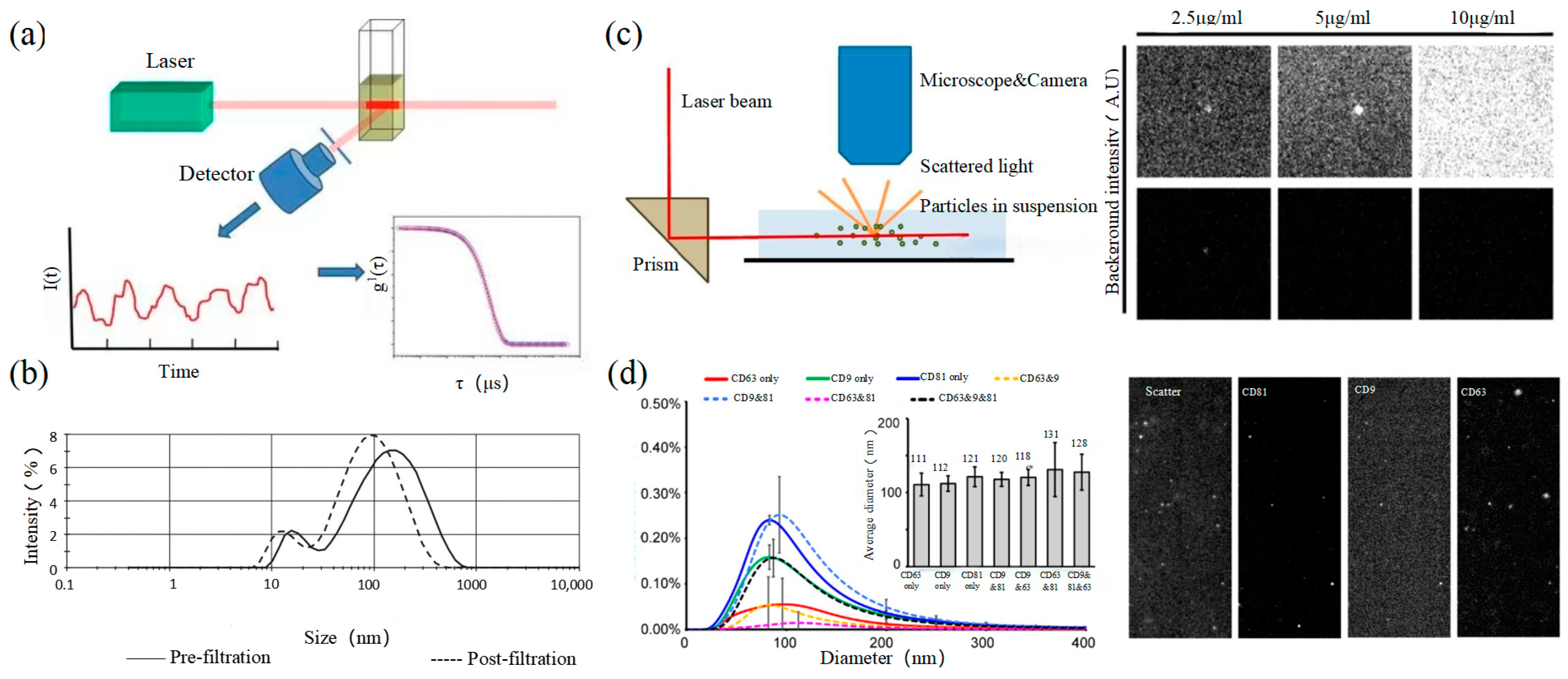
2.3. Dynamic Light Scattering
Dynamic light scattering (DLS) is an optical analysis method for measuring the size and distribution of sub-micron particles, and its basic principle is shown in Figure 3a. The Brownian motion of the particles causes the change in the light scattering signal, which is monitored by a digital autocorrelator to calculate the diffusion velocity and particle distribution of the particles [62]. The motion rate of small particles is higher, and its intensity fluctuation is larger. As a result, there is a swift and pronounced decline in the correlation curve. The sensitivity and FOV of DLS are better than the above papers, which can realize the basic characterization of large-size exosome nanoparticles. For smaller particles, the behavior of self-polymerization interferes with the light-intensity signal. Thus, it is impossible to realize the precision analysis and detection of high-concentration samples. In 2009, Lawrie et al. used DLS to characterize the size distribution of EV derived from red blood cells [63]. DLS analyzes all particles in a sample simultaneously, and therefore the information on the number or concentration of a certain category of particle cannot be provided [64]. For example, DLS provides a clear range of the diameter of EVs derived from ovarian cancer cells, but the concentration is difficult to analyze [65]. Therefore, DLS technology is often combined with other technologies to complete the characterization of EVs. For example, Tajik T combined DLS technology with electron microscopy, showed cannabis-derived EVs (CDEVs) can be considered exosome-like nanovesicles, and highlighted that CDEVs can be an ideal natural vehicle for bioactive phytocannabinoids, promoting the research of EVs in cancer diagnosis [66].
2.4. Nanoparticle Tracking Analysis
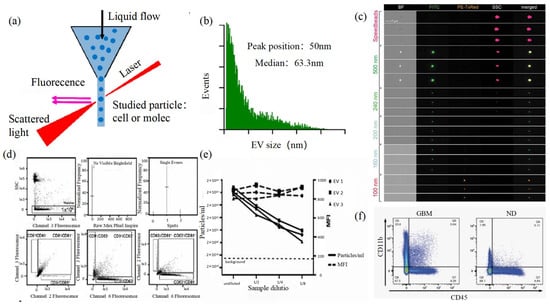
2.5. Flow Cytometry
3. Super-Resolution Imaging Technology
3.1. Single Molecule Localization Imaging Technology
3.1.1. Stochastic Optical Reconstruction Microscopy and Photoactivated Localization Microscopy Technology
3.1.2. DNA-PAINT Technology
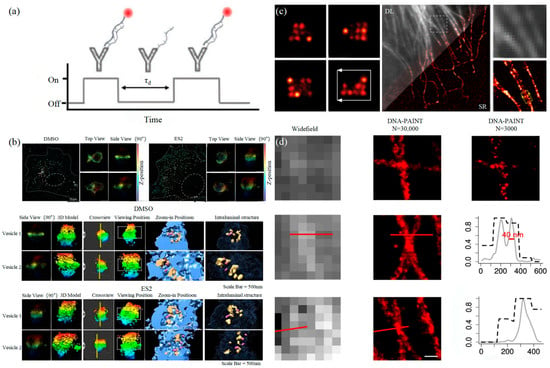
3.2. Stimulated Emission Depletion Technology
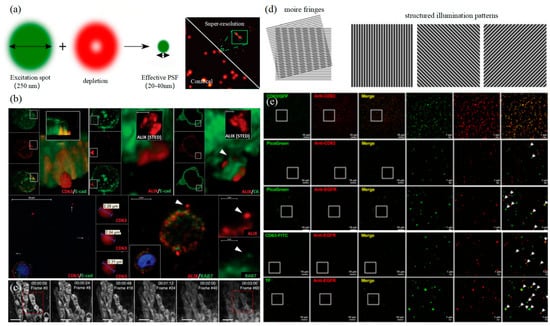
3.3. Structured Illumination Microscopy Technology
4. Summarize the Outlook
References
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238.
- Alix-Panabières, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118.
- Guo, L.C.; He, B. Extracellular vesicles and their diagnostic and prognostic potential in cancer. Transl. Cancer Res. 2017, 6, 599–612.
- Huang, T.; Deng, C.X. Current Progresses of Exosomes as Cancer Diagnostic and Prognostic Biomarkers. Int. J. Biol. Sci. 2019, 15, 1–11.
- Im, E.J.; Lee, C.H.; Moon, P.G.; Rangaswamy, G.G.; Lee, B.; Lee, J.M.; Lee, J.C.; Jee, J.G.; Bae, J.S.; Kwon, T.K.; et al. Sulfisoxazole inhibits the secretion of small extracellular vesicles by targeting the endothelin receptor A. Nat. Commun. 2019, 10, 1387.
- Junker, K.; Heinzelmann, J.; Beckham, C.; Ochiya, T.; Jenster, G. Extracellular Vesicles and Their Role in Urologic Malignancies. Eur. Urol. 2016, 70, 323–331.
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Phillips, S.; Kaimal, V.; Mao, Y.; Hua, W.; Yang, I.; Fu, C.C.; Nolan, J.; et al. miRNA contents of cerebrospinal fluid extracellular vesicles in glioblastoma patients. J. Neuro-Oncol. 2015, 123, 205–216.
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51.
- Ciardiello, C.; Cavallini, L.; Spinelli, C.; Yang, J.; Reis-Sobreiro, M.; de Candia, P.; Minciacchi, V.R.; Di Vizio, D. Focus on Extracellular Vesicles: New Frontiers of Cell-to-Cell Communication in Cancer. Int. J. Mol. Sci. 2016, 17, 175.
- Liu, J.; Ren, L.; Li, S.; Li, W.; Zheng, X.; Yang, Y.; Fu, W.; Yi, J.; Wang, J.; Du, G. The biology, function, and applications of exosomes in cancer. Acta Pharm. Sin. B 2021, 11, 2783–2797.
- Duijvesz, D.; Luider, T.; Bangma, C.H.; Jenster, G. Exosomes as biomarker treasure chests for prostate cancer. Eur. Urol. 2011, 59, 823–831.
- Kimiz-Gebologlu, I.; Oncel, S.S. Exosomes: Large-scale production, isolation, drug loading efficiency, and biodistribution and uptake. J. Control. Release 2022, 347, 533–543.
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442.
- Berditchevski, F.; Odintsova, E. Tetraspanins as regulators of protein trafficking. Traffic 2007, 8, 89–96.
- Rezaie, J.; Akbari, A.; Rahbarghazi, R. Inhibition of extracellular vesicle biogenesis in tumor cells: A possible way to reduce tumorigenesis. Cell Biochem. Funct. 2022, 40, 248–262.
- Boucheix, C.; Rubinstein, E. Tetraspanins. Cell. Mol. Life Sci. 2001, 58, 1189–1205.
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255.
- He, M.; Crow, J.; Roth, M.; Zeng, Y.; Godwin, A.K. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip 2014, 14, 3773–3780.
- Nikishin, I.; Dulimov, R.; Skryabin, G.; Galetsky, S.; Tchevkina, E.; Bagrov, D. ScanEV—A neural network-based tool for the automated detection of extracellular vesicles in TEM images. Micron 2021, 145, 103044.
- Kahlert, C.; Kalluri, R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 2013, 91, 431–437.
- Wang, X.; Huang, J.; Chen, W.; Li, G.; Li, Z.; Lei, J. The updated role of exosomal proteins in the diagnosis, prognosis, and treatment of cancer. Exp. Mol. Med. 2022, 54, 1390–1400.
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.
- Risha, Y.; Minic, Z.; Ghobadloo, S.M.; Berezovski, M.V. The proteomic analysis of breast cell line exosomes reveals disease patterns and potential biomarkers. Sci. Rep. 2020, 10, 13572.
- Haraszti, R.A.; Didiot, M.C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; DiFiglia, M.; Kiebish, M.A.; et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 2016, 5, 32570.
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797.
- Yang, B.; Chen, Y.; Shi, J. Exosome Biochemistry and Advanced Nanotechnology for Next-Generation Theranostic Platforms. Adv. Mater. 2019, 31, e1802896.
- Yu, D.; Li, Y.; Wang, M.; Gu, J.; Xu, W.; Cai, H.; Fang, X.; Zhang, X. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer 2022, 21, 56.
- Hsu, M.T.; Wang, Y.K.; Tseng, Y.J. Exosomal Proteins and Lipids as Potential Biomarkers for Lung Cancer Diagnosis, Prognosis, and Treatment. Cancers 2022, 14, 732.
- Lee, Y.R.; Kim, G.; Tak, W.Y.; Jang, S.Y.; Kweon, Y.O.; Park, J.G.; Lee, H.W.; Han, Y.S.; Chun, J.M.; Park, S.Y.; et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int. J. Cancer 2019, 144, 1444–1452.
- Zhu, L.; Sun, H.T.; Wang, S.; Huang, S.L.; Zheng, Y.; Wang, C.Q.; Hu, B.Y.; Qin, W.; Zou, T.T.; Fu, Y.; et al. Isolation and characterization of exosomes for cancer research. J. Hematol. Oncol. 2020, 13, 152.
- Liu, Y.; Zhou, Z.; Wang, F.; Kewes, G.; Wen, S.; Burger, S.; Ebrahimi Wakiani, M.; Xi, P.; Yang, J.; Yang, X.; et al. Axial localization and tracking of self-interference nanoparticles by lateral point spread functions. Nat. Commun. 2021, 12, 2019.
- Wu, H.J.; Huang, C.L.; Wang, L.W.; Li, Q.H.; Li, Y.J.; Zhang, L.H.; Zhu, D.W. Folate-targeted co-delivery polymersomes for efficient photo-chemo-antiangiogenic therapy against breast cancer and in vivo evaluation via OCTA_NIRF dual-modal imaging-3. Chin. Chem. Lett. 2022, 33, 5035–5041.
- Filipe, V.; Hawe, A.; Jiskoot, W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm. Res. 2010, 27, 796–810.
- Libregts, S.; Arkesteijn, G.J.A.; Németh, A.; Nolte-'t Hoen, E.N.M.; Wauben, M.H.M. Flow cytometric analysis of extracellular vesicle subsets in plasma: Impact of swarm by particles of non-interest. J. Thromb. Haemost. 2018, 16, 1423–1436.
- Arraud, N.; Linares, R.; Tan, S.; Gounou, C.; Pasquet, J.M.; Mornet, S.; Brisson, A.R. Extracellular vesicles from blood plasma: Determination of their morphology, size, phenotype and concentration. J. Thromb. Haemost. 2014, 12, 614–627.
- Lai, J.J.; Chau, Z.L.; Chen, S.Y.; Hill, J.J.; Korpany, K.V.; Liang, N.W.; Lin, L.H.; Lin, Y.H.; Liu, J.K.; Liu, Y.C.; et al. Exosome Processing and Characterization Approaches for Research and Technology Development. Adv. Sci. 2022, 9, e2103222.
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75.
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145.
- Jiang, C.; Zhang, N.; Hu, X.; Wang, H. Tumor-associated exosomes promote lung cancer metastasis through multiple mechanisms. Mol. Cancer 2021, 20, 117.
- Zhao, S.; Mi, Y.; Guan, B.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Cai, S.; Xu, Y.; Li, X.; et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 2020, 13, 156.
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. (BBA) Acta Rev. Cancer 2019, 1871, 455–468.
- Hartjes, T.A.; Mytnyk, S.; Jenster, G.W.; van Steijn, V.; van Royen, M.E. Extracellular Vesicle Quantification and Characterization: Common Methods and Emerging Approaches. Bioengineering 2019, 6, 7.
- Maas, S.L.; De Vrij, J.; Broekman, M.L. Quantification and size-profiling of extracellular vesicles using tunable resistive pulse sensing. J. Vis. Exp. 2014, 92, e51623.
- Maas, S.L.; Broekman, M.L.; de Vrij, J. Tunable Resistive Pulse Sensing for the Characterization of Extracellular Vesicles. Methods Mol. Biol. 2017, 1545, 21–33.
- Durak-Kozica, M.; Baster, Z.; Kubat, K.; Stępień, E. 3D visualization of extracellular vesicle uptake by endothelial cells. Cell. Mol. Biol. Lett. 2018, 23, 57.
- Vogel, R.; Pal, A.K.; Jambhrunkar, S.; Patel, P.; Thakur, S.S.; Reátegui, E.; Parekh, H.S.; Saá, P.; Stassinopoulos, A.; Broom, M.F. High-Resolution Single Particle Zeta Potential Characterisation of Biological Nanoparticles using Tunable Resistive Pulse Sensing. Sci. Rep. 2017, 7, 17479.
- Malenica, M.; Vukomanović, M.; Kurtjak, M.; Masciotti, V.; Dal Zilio, S.; Greco, S.; Lazzarino, M.; Krušić, V.; Perčić, M.; Jelovica Badovinac, I.; et al. Perspectives of Microscopy Methods for Morphology Characterisation of Extracellular Vesicles from Human Biofluids. Biomedicines 2021, 9, 603.
- McNamara, R.P.; Zhou, Y.; Eason, A.B.; Landis, J.T.; Chambers, M.G.; Willcox, S.; Peterson, T.A.; Schouest, B.; Maness, N.J.; MacLean, A.G.; et al. Imaging of surface microdomains on individual extracellular vesicles in 3-D. J. Extracell. Vesicles 2022, 11, e12191.
- Fakhredini, F.; Mansouri, E.; Mard, S.A.; Valizadeh Gorji, A.; Rashno, M.; Orazizadeh, M. Effects of Exosomes Derived from Kidney Tubular Cells on Diabetic Nephropathy in Rats. Cell J. 2022, 24, 28–35.
- Conde-Vancells, J.; Rodriguez-Suarez, E.; Embade, N.; Gil, D.; Matthiesen, R.; Valle, M.; Elortza, F.; Lu, S.C.; Mato, J.M.; Falcon-Perez, J.M. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 2008, 7, 5157–5166.
- Jeong, D.; Kim, M.J.; Park, Y.; Chung, J.; Kweon, H.S.; Kang, N.G.; Hwang, S.J.; Youn, S.H.; Hwang, B.K.; Kim, D. Visualizing extracellular vesicle biogenesis in gram-positive bacteria using super-resolution microscopy. BMC Biol. 2022, 20, 270.
- Zabeo, D.; Cvjetkovic, A.; Lässer, C.; Schorb, M.; Lötvall, J.; Höög, J.L. Exosomes purified from a single cell type have diverse morphology. J. Extracell. Vesicles 2017, 6, 1329476.
- Gardiner, C.; Di Vizio, D.; Sahoo, S.; Théry, C.; Witwer, K.W.; Wauben, M.; Hill, A.F. Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. J. Extracell. Vesicles 2016, 5, 32945.
- Issman, L.; Brenner, B.; Talmon, Y.; Aharon, A. Cryogenic transmission electron microscopy nanostructural study of shed microparticles. PLoS ONE 2013, 8, e83680.
- Jung, M.K.; Mun, J.Y. Sample Preparation and Imaging of Exosomes by Transmission Electron Microscopy. J. Vis. Exp. 2018, 131, e56482.
- Božič, D.; Hočevar, M.; Kisovec, M.; Pajnič, M.; Pađen, L.; Jeran, M.; Bedina Zavec, A.; Podobnik, M.; Kogej, K.; Iglič, A.; et al. Stability of Erythrocyte-Derived Nanovesicles Assessed by Light Scattering and Electron Microscopy. Int. J. Mol. Sci. 2021, 22, 12772.
- Sokolova, V.; Ludwig, A.K.; Hornung, S.; Rotan, O.; Horn, P.A.; Epple, M.; Giebel, B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf. B Biointerfaces 2011, 87, 146–150.
- Cizmar, P.; Yuana, Y. Detection and Characterization of Extracellular Vesicles by Transmission and Cryo-Transmission Electron Microscopy. Methods Mol. Biol. 2017, 1660, 221–232.
- Liu, Z.; Xue, H.; Chen, Q.; Yang, G. A method for extraction of exosomes from breast tumour cells and characterisation by transmission electron microscopy. J. Microsc. 2023, 292, 117–122.
- Park, Y.H.; Shin, H.W.; Jung, A.R.; Kwon, O.S.; Choi, Y.J.; Park, J.; Lee, J.Y. Prostate-specific extracellular vesicles as a novel biomarker in human prostate cancer. Sci. Rep. 2016, 6, 30386.
- Kurtjak, M.; Kereïche, S.; Klepac, D.; Križan, H.; Perčić, M.; Krušić Alić, V.; Lavrin, T.; Lenassi, M.; Wechtersbach, K.; Kojc, N.; et al. Unveiling the Native Morphology of Extracellular Vesicles from Human Cerebrospinal Fluid by Atomic Force and Cryogenic Electron Microscopy. Biomedicines 2022, 10, 1251.
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427.
- Palmieri, V.; Lucchetti, D.; Gatto, I.; Maiorana, A.; Marcantoni, M.; Maulucci, G.; Papi, M.; Pola, R.; De Spirito, M.; Sgambato, A. Dynamic light scattering for the characterization and counting of extracellular vesicles: A powerful noninvasive tool. J. Nanopart. Res. 2014, 16, 2583.
- Perpetuo, L.; Ferreira, R.; Thongboonkerd, V.; Guedes, S.; Amado, F.; Vitorino, R. Urinary exosomes: Diagnostic impact with a bioinformatic approach. Adv. Clin. Chem. 2022, 111, 69–99.
- Gercel-Taylor, C.; Atay, S.; Tullis, R.H.; Kesimer, M.; Taylor, D.D. Nanoparticle analysis of circulating cell-derived vesicles in ovarian cancer patients. Anal. Biochem. 2012, 428, 44–53.
- Tajik, T.; Baghaei, K.; Moghadam, V.E.; Farrokhi, N.; Salami, S.A. Extracellular vesicles of cannabis with high CBD content induce anticancer signaling in human hepatocellular carcinoma. Biomed. Pharmacother. 2022, 152, 113209.
- Hassan, P.A.; Rana, S.; Verma, G. Making sense of Brownian motion: Colloid characterization by dynamic light scattering. Langmuir 2015, 31, 3–12.
- Lawrie, A.S.; Albanyan, A.; Cardigan, R.A.; Mackie, I.J.; Harrison, P. Microparticle sizing by dynamic light scattering in fresh-frozen plasma. Vox Sang. 2009, 96, 206–212.
- Szatanek, R.; Baj-Krzyworzeka, M.; Zimoch, J.; Lekka, M.; Siedlar, M.; Baran, J. The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int. J. Mol. Sci. 2017, 18, 1153.
- Cho, S.; Yi, J.; Kwon, Y.; Kang, H.; Han, C.; Park, J. Multifluorescence Single Extracellular Vesicle Analysis by Time-Sequential Illumination and Tracking. ACS Nano 2021, 15, 11753–11761.
- Vestad, B.; Llorente, A.; Neurauter, A.; Phuyal, S.; Kierulf, B.; Kierulf, P.; Skotland, T.; Sandvig, K.; Haug, K.B.F.; Øvstebø, R. Size and concentration analyses of extracellular vesicles by nanoparticle tracking analysis: A variation study. J. Extracell. Vesicles 2017, 6, 1344087.
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.; Hole, P.; Carr, B.; Redman, C.W.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 780–788.
- van der Pol, E.; Coumans, F.A.; Grootemaat, A.E.; Gardiner, C.; Sargent, I.L.; Harrison, P.; Sturk, A.; van Leeuwen, T.G.; Nieuwland, R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J. Thromb. Haemost. 2014, 12, 1182–1192.
- Hu, Y.; Tian, Y.; Di, H.; Xue, C.; Zheng, Y.; Hu, B.; Lin, Q.; Yan, X. Noninvasive Diagnosis of Nasopharyngeal Carcinoma Based on Phenotypic Profiling of Viral and Tumor Markers on Plasma Extracellular Vesicles. Anal. Chem. 2022, 94, 9740–9749.
- Liu, H.S.; Tian, Y.; Xue, C.F.; Niu, Q.; Chen, C.; Yan, X.M. Analysis of extracellular vesicle DNA at the single-vesicle level by nano-flow cytometry. J. Extracell. Vesicles 2022, 11, e12206.
- Ricklefs, F.L.; Maire, C.L.; Reimer, R.; Dührsen, L.; Kolbe, K.; Holz, M.; Schneider, E.; Rissiek, A.; Babayan, A.; Hille, C.; et al. Imaging flow cytometry facilitates multiparametric characterization of extracellular vesicles in malignant brain tumours. J. Extracell. Vesicles 2019, 8, 1588555.
- Aibaidula, A.Z.; Fain, C.E.; Garcia, L.C.; Wier, A.; Bouchal, S.M.; Bauman, M.M.; Jung, M.Y.; Sarkaria, J.N.; Johnson, A.J.; Parney, I.F. Spectral flow cytometry identifies distinct nonneoplastic plasma extracellular vesicle phenotype in glioblastoma patients. Neuro-Oncol. Adv. 2023, 5, vdad082.
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645.
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–795.
- Jones, S.A.; Shim, S.H.; He, J.; Zhuang, X. Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods 2011, 8, 499–508.
- Shim, S.H.; Xia, C.; Zhong, G.; Babcock, H.P.; Vaughan, J.C.; Huang, B.; Wang, X.; Xu, C.; Bi, G.Q.; Zhuang, X. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad. Sci. USA 2012, 109, 13978–13983.
- Zong, S.; Zong, J.; Chen, C.; Jiang, X.; Zhang, Y.; Wang, Z.; Cui, Y. Single molecule localization imaging of exosomes using blinking silicon quantum dots. Nanotechnology 2018, 29, 065705.
- Jungmann, R.; Avendaño, M.S.; Woehrstein, J.B.; Dai, M.; Shih, W.M.; Yin, P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 2014, 11, 313–318.
- Tholen, M.M.E.; Tas, R.P.; Wang, Y.; Albertazzi, L. Beyond DNA: New probes for PAINT super-resolution microscopy. Chem. Commun. 2023, 59, 8332–8342.
- Chen, C.; Zong, S.; Liu, Y.; Wang, Z.; Zhang, Y.; Chen, B.; Cui, Y. Profiling of Exosomal Biomarkers for Accurate Cancer Identification: Combining DNA-PAINT with Machine- Learning-Based Classification. Small 2019, 15, e1901014.
- Liu, D.A.; Tao, K.; Wu, B.; Yu, Z.; Szczepaniak, M.; Rames, M.; Yang, C.; Svitkina, T.; Zhu, Y.; Xu, F.; et al. A phosphoinositide switch mediates exocyst recruitment to multivesicular endosomes for exosome secretion. Nat. Commun. 2023, 14, 6883.
- Auer, A.; Strauss, M.T.; Schlichthaerle, T.; Jungmann, R. Fast, Background-Free DNA-PAINT Imaging Using FRET-Based Probes. Nano Lett. 2017, 17, 6428–6434.
- Zhu, M.; Zhang, L.; Jin, L.; Chen, J.; Zhang, Y.; Xu, Y. DNA-PAINT Imaging Accelerated by Machine Learning. Front. Chem. 2022, 10, 864701.
- van Wee, R.; Filius, M.; Joo, C. Completing the canvas: Advances and challenges for DNA-PAINT super-resolution imaging. Trends Biochem. Sci. 2021, 46, 918–930.
- McEvoy, A.L.; Greenfield, D.; Bates, M.; Liphardt, J. Q&A: Single-molecule localization microscopy for biological imaging. BMC Biol. 2010, 8, 106.
- Nieves, D.J.; Gaus, K.; Baker, M.A.B. DNA-Based Super-Resolution Microscopy: DNA-PAINT. Genes 2018, 9, 621.
- Li, W.; Li, C.; Zhou, T.; Liu, X.; Liu, X.; Li, X.; Chen, D. Role of exosomal proteins in cancer diagnosis. Mol. Cancer 2017, 16, 145.
- Hell, S.W.; Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994, 19, 780–782.
- Sezgin, E. Super-resolution optical microscopy for studying membrane structure and dynamics. J. Phys. Condens. Matter 2017, 29, 273001.
- Huang, G.; Liu, Y.; Wang, D.; Zhu, Y.; Wen, S.; Ruan, J.; Jin, D. Upconversion nanoparticles for super-resolution quantification of single small extracellular vesicles. eLight 2022, 2, 20.
- Valcz, G.; Buzás, E.I.; Kittel, Á.; Krenács, T.; Visnovitz, T.; Spisák, S.; Török, G.; Homolya, L.; Zsigrai, S.; Kiszler, G.; et al. En bloc release of MVB-like small extracellular vesicle clusters by colorectal carcinoma cells. J. Extracell. Vesicles 2019, 8, 1596668.
- Yang, X.; Yang, Z.; Wu, Z.; He, Y.; Shan, C.; Chai, P.; Ma, C.; Tian, M.; Teng, J.; Jin, D.; et al. Mitochondrial dynamics quantitatively revealed by STED nanoscopy with an enhanced squaraine variant probe. Nat. Commun. 2020, 11, 3699.
- Choi, D.; Montermini, L.; Jeong, H.; Sharma, S.; Meehan, B.; Rak, J. Mapping Subpopulations of Cancer Cell-Derived Extracellular Vesicles and Particles by Nano-Flow Cytometry. ACS Nano 2019, 13, 10499–10511.
- Gustafsson, M.G. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 2000, 198, 82–87.
- Gustafsson, M.G. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc. Natl. Acad. Sci. USA 2005, 102, 13081–13086.
- Li, D.; Shao, L.; Chen, B.C.; Zhang, X.; Zhang, M.; Moses, B.; Milkie, D.E.; Beach, J.R.; Hammer, J.A., 3rd; Pasham, M.; et al. ADVANCED IMAGING. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 2015, 349, aab3500.
- Boland, M.A.; Cohen, E.A.K.; Flaxman, S.R.; Neil, M.A.A. Improving axial resolution in Structured Illumination Microscopy using deep learning. Philos. Trans. Ser. A Math. Phys. Eng. Sci. 2021, 379, 20200298.
- He, Y.; Yao, Y.; He, Y.; Huang, Z.; Luo, F.; Zhang, C.; Qi, D.; Jia, T.; Wang, Z.; Sun, Z.; et al. Surpassing the resolution limitation of structured illumination microscopy by an untrained neural network. Biomed. Opt. Express 2023, 14, 106–117.
- Butola, A.; Acuna, S.; Hansen, D.H.; Agarwal, K. Scalable-resolution structured illumination microscopy. Opt. Express 2022, 30, 43752–43767.




