Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Peter Matúš | -- | 1588 | 2023-06-21 10:40:57 | | | |
| 2 | Rita Xu | Meta information modification | 1588 | 2023-06-21 10:56:43 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Balíková, K.; Vojtková, H.; Duborská, E.; Kim, H.; Matúš, P.; Urík, M. Biosynthesis of Extracellular Polysaccharides in Pseudomonas. Encyclopedia. Available online: https://encyclopedia.pub/entry/45912 (accessed on 08 February 2026).
Balíková K, Vojtková H, Duborská E, Kim H, Matúš P, Urík M. Biosynthesis of Extracellular Polysaccharides in Pseudomonas. Encyclopedia. Available at: https://encyclopedia.pub/entry/45912. Accessed February 08, 2026.
Balíková, Katarína, Hana Vojtková, Eva Duborská, Hyunjung Kim, Peter Matúš, Martin Urík. "Biosynthesis of Extracellular Polysaccharides in Pseudomonas" Encyclopedia, https://encyclopedia.pub/entry/45912 (accessed February 08, 2026).
Balíková, K., Vojtková, H., Duborská, E., Kim, H., Matúš, P., & Urík, M. (2023, June 21). Biosynthesis of Extracellular Polysaccharides in Pseudomonas. In Encyclopedia. https://encyclopedia.pub/entry/45912
Balíková, Katarína, et al. "Biosynthesis of Extracellular Polysaccharides in Pseudomonas." Encyclopedia. Web. 21 June, 2023.
Copy Citation
Pseudomonas biofilms have been studied intensively for several decades and research outcomes have been successfully implemented in various medical and agricultural applications. Research on biofilm synthesis and composition has also overlapped with the objectives of environmental sciences, since biofilm components show exceptional physicochemical properties applicable to remediation techniques. Especially, exopolysaccharides (ExPs) have been at the center of scientific interest, indicating their potential in solving the environmental issues of heavy metal land and water contamination via sorptive interactions and flocculation.
exopolysaccharides
Pseudomonas
biosorption
bioremediation
1. Introduction
Bacterial exopolysaccharides (ExPs), a group of extracellular polymeric substances (EPSs), are the structural and functional components of microbial biofilms that display exceptional physicochemical properties. Thus, bacterial ExPs with unique attributes have found their way into biomedical science practice (e.g., tissue engineering) and have been successfully implemented into a myriad of industrial and medical applications [1][2].
Interest in ExP-producing bacteria has also expanded into the research areas of environmental sciences, including studies on eco-friendly municipal and wastewater treatment processes [3]. This is because many standard remediation techniques require the usage of reagents that may be hard to degrade, or various environmentally harmful by-products are produced during their utilization at contaminated sites. This includes chemical treatment, which usually detoxifies metals via redox transformation or neutralization by application of the reagents, such as potassium permanganate, hydrogen peroxide, hypochlorite, synthetic surfactants, or chlorine gas, to precipitate, immobilize, or preconcentrate the hazardous contaminants [4][5][6]. The chemical leaching of soils and sediments by applying strong inorganic and organic acids and persistent synthetic chelating agents (e.g., ethylenediaminetetraacetic acid or its derivatives) to solubilize contaminants has also been successfully tested for heavy metal removal [7]. Reactive solid inorganic and biological substances, as well as materials with active surfaces (e.g., zero-valent iron, ferric oxides and oxohydroxides, nanomaterials, zeolite, biological waste) have been studied as potential components of permeable treatment barriers to restrict the movement of the contaminant in the environment [8][9][10][11]. However, in some cases, unpredictable effects regarding the toxicity and mobility of generated species can be expected since these interactions are usually non-specific. The application of electrochemical and electrokinetic remediation methods, engineered to site-specific requirements, has been performed for heavy metal removal [12], showing promising results in combination with other remediation approaches, including novel biochemical methods [13]. More prominent green approaches include phytoremediation, phytoextraction, and biosorption. They are usually performed in conjunction with other methods, e.g., chemical leaching [14]. Still, the application of microbial ExPs is environmentally advantageous since these biogenic polymers are usually water soluble, susceptible to natural degradation, and less harmful than synthetic polymers [15].
Bacterial ExPs find their successful applications in heavy metal removal, oil recovery, and various in situ remediation techniques such as emulsifiers, sorbents, biofilters, surfactants, and bioflocculants [16][17][18]. The interest of environmental researchers in ExP-producing bacteria is also highlighted in several patent deposits focusing on the prosperous application of bacteria in the remediation of contaminated sites. Villela et al. [19] reported that there are 114 patents describing the degradation of oil compounds exclusively by Pseudomonas, thus highlighting the leading role of this bacterial genus in hydrocarbon-contaminated site remediation.
The utilization of Pseudomonas strains in remediation is not limited solely to the biodegradation of hydrocarbons; they have also been successfully applied for the decontamination of heavy metal-polluted waters, soils, and sediments [20]. This is primarily due to their ability to produce metal-chelating siderophores and surface-active extracellular polymeric substances [21]. Regarding the latter, Pseudomonas species are considered high-ExP-producing organisms [22], and since they are ubiquitous, being isolated from various types of environments, including industrial waste and activated sludge [23], they are considered potent in solving the issue of heavy metal contamination [24].
2. Biosynthesis of Extracellular Polysaccharides in Pseudomonas
Extensive progress has been made in elucidating the synthesis of bacterial extracellular homopolysaccharides and heteropolysaccharides in recent years (Figure 1). They are synthesized by bacteria either extracellularly (outside the cell membrane and the cell wall), within the cell wall, or intracellularly [25].
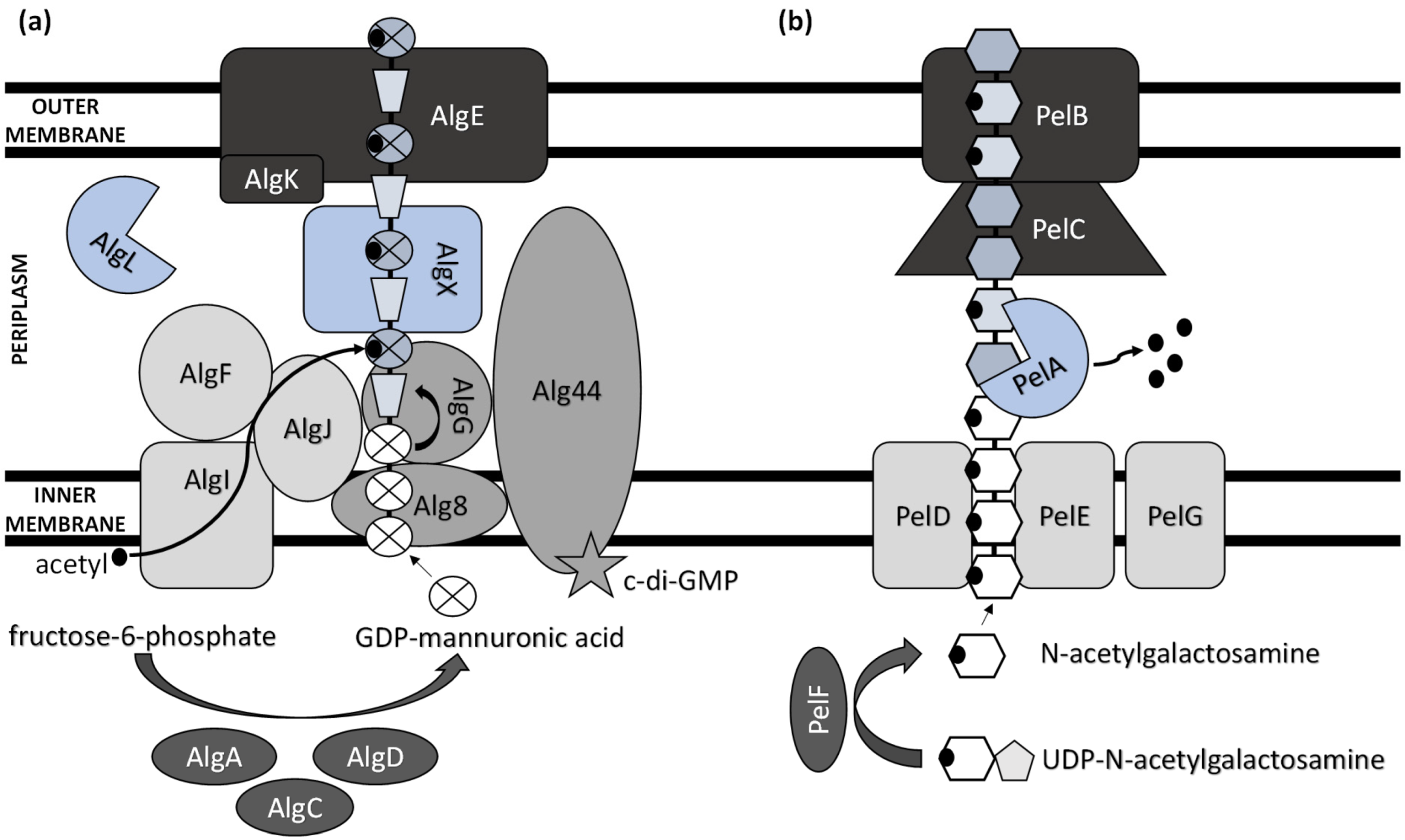
Figure 1. Schematic model of (a) alginate and (b) Pel polysaccharide biosynthetic machinery.
While heteropolysaccharides are mostly synthesized intracellularly and transported outside the cell, homopolysaccharide production generally involves the activity of enzymes secreted by the bacterium to the extracellular environment. ExP production comprises several steps, including the synthesis of ExP precursors, repeat-unit assembly on a lipid carrier located at the cytoplasmic membrane, modification (e.g., acylation, acetylation, sulphation, and methylation), membrane translocation, polymerization, and export [26]. Thus, there are several functionally distinguished enzymes required for ExPs’ synthesis and development [27]. Several other enzymes, which are not unique to ExP production and serve as intermediates in protein regulation and central carbon metabolism, are also involved in ExPs’ biosynthesis process [28].
The biosynthesis of ExPs requires the involvement of activated monosaccharides derived from catabolized sugars. These include sugar nucleosides (nucleoside diphosphate sugars) or their derivatives (e.g., uridine diphosphate (UDP)-N-acetylglucosamine and guanosine diphosphate (GDP)-mannuronic acid) [29].
The main ExP component in the bacteria of Pseudomonas genera is alginate, an anionic linear polymer composed of beta-1,4-linked mannuronic acids (M-blocks) and C5-epimer α-L-guluronic acid (G-blocks) [30]. Alginate’s viscosifying, gelling, and stabilizing properties make this biopolymer an important industrial polysaccharide with great prospects in applications such as drug and protein delivery systems and food encapsulation [31][32]. Thirteen proteins are directly involved in the biosynthesis of alginate, and except for AlgC, they are all encoded by the alg operon [33]. The alginate precursor is synthesized by three enzymes including AlgA (bifunctional enzyme phosphomannose isomerase/guanosine 5′-diphospho-D-mannose pyrophosphorylase), AlgC (phosphomannomutase) and AlgD (GDP-mannose dehydrogenase), which allow the conversion of fructose-6-phosphate to GDP-mannuronic acid via four steps [34][35][36]. Alginate is first synthesized as a linear homopolymer from the GDP-mannuronic acid to polymannuronic acid by catalytic subunit Alg8 (alpha-1,3-glucosyltransferase) which interacts with Alg44 co-polymerase located at the cytoplasmatic membrane [37], with most of the latter being exposed to the periplasm [38]. These enzymes allowed the movement of the alginate precursor across the inner membrane for polymerization [39]. Alg44 also demonstrated the capability of binding the second messenger cyclic dimeric guanosine monophosphate (c-di-GMP) synthesized by MucR, a membrane-anchored protein, which is required for alginate biosynthesis [40].
In the periplasm, the polymannuronate is modified by epimerization or acetylation. AlgI, AlgJ, and AlgF are required for the addition of O-acetyl groups to the alginate polymer at O2 and/or O3 positions, which is an essential process for the stabilization of the intracellular alginate matrix for microcolony formation [41]. Acetylation can also affect epimerization reactions, since the activity of AlgG, a C5-epimerase that directly converts D-mannuronate to L-guluronate, has been detected only when the acetyl groups are removed from the polymannuronate substrate [42]. Newly formed macromolecules are most likely transported within the periplasm by the periplasmic protein AlgX that surrounds and protects the polymers from degradation by AlgL, a periplasmic alginate lyase [43][44]. Alginate is then secreted by the putative export protein AlgE [45]. The proper localization of AlgE for the periplasmic components of the alginate’s biosynthetic machinery is facilitated by AlgK [46].
The acid hydrolysis method is usually applied for the determination of ExPs’ monomeric components (e.g., glucose, fructose, galactose, and arabinose). Myszka and Czaczyk [47] reported that, under starvation conditions (ABPG medium reduced by 90% (w/v) of optimal nutrient availability), the EPS matrix of Pseudomonas aeruginosa ATCC 10145 consisted solely of glucosyl units. Grob, et al. [48] suggested that the survival of P. aeruginosa SG41 under highly chlorinated conditions was enabled by the overproduction of alginate, a major component of the SG41 strain’s ExPs (109.8 µg·g−1 cell dry mass) [49]. Thus, alginate overproduction is advantageous in harsh environments. Still, the nonmucoid P. aeruginosa strains that are the predominant environmental phenotype do not need to express the alginate biosynthetic genes to form the nonmucoid biofilms [50]. These use either Pel or Psl as the primary matrix structural polysaccharide [51].
A previously reported analysis of Pel polymer suggested that it is rich in cationic amino sugars, N-acetylgalactosamine, and N-acetylglucosamine, in a 5:1 ratio [52]. However, just recently, Le Mauff et al. [53] characterized the configuration and structure of Pel and suggested that it is a polymer of partially de-N-acetylated α-1,4-N-acetylgalactosamine, and it does not contain N-acetylglucosamine.
Pel synthesis requires protein products of a seven-gene operon pelABCDEFG [54]. The protein complex of PelD, PelE, PelF, and PelG is very likely a Pel-polysaccharide synthase whose activity is dependent on the localization of cytosolic glycosyltransferase PelF [55] to the inner membrane protein complex PelDEG [56]. PelDEG is also competent for the transport of the Pel polymer across the cytoplasmatic membrane.
PelA is a multi-domain protein that localizes to both the periplasm and membrane and exhibits both hydrolase and de-N-acetylase activity [57][58]. The PelBC complex is responsible for the transport of the matured polymer into the extracellular milieu. PelB is located at the outer membrane and contains a transmembrane β-barrel porin towards which the lipoprotein PelC, which is localized to the inner leaflet of the outer membrane, guides the positively charged Pel and, thus, acts as a charged molecular funnel facilitating Pel export [59].
Psl is a neutral branched pentasaccharide comprising D-mannose, D-glucose, and L-rhamnose. It is synthesized by the polysaccharide synthesis locus (psl). The psl gene cluster consists of 15 genes, of which 11 are necessary for Psl polysaccharide synthesis (pslACDEFGHIJKL) [60]. However, the specific function of each protein is not completely understood.
pslB likely encodes a bifunctional enzyme with GDP-mannose pyrophosphorylase/phosphomannose isomerase dual activities, which is the only enzyme from the psl operon involved in sugar nucleotide precursor production [61]. The inner membrane-associated glycoside hydrolase PslG can be involved in the biosynthesis of Psl polysaccharide [60], although its role in this process is controversial [62]. Since the five PslAEJKL proteins have inner membrane-spanning domains, it was hypothesized that they make up the Psl polymerization complex [33]. Regarding Psl translocation and export, the complex of PslD and PslE helps transport Psl across the outer membrane [63].
References
- Mohd Nadzir, M.; Nurhayati, R.W.; Idris, F.N. Biomedical applications of bacterial exopolysaccharides: A review. Polymers 2021, 13, 530.
- Nwodo, U.U.; Green, E.; Okoh, A.I. Bacterial exopolysaccharides: Functionality and prospects. Int. J. Mol. Sci. 2012, 13, 14002–14015.
- Kanmani, P.; Yuvapriya, S. Exopolysaccharide from Bacillus sp. YP03: Its properties and application as a flocculating agent in wastewater treatment. Int. J. Environ. Sci. Technol. 2018, 15, 2551–2560.
- Mulligan, C.N.; Yong, R.N.; Gibbs, B.F. Remediation technologies for metal-contaminated soils and groundwater: An evaluation. Eng. Geol. 2001, 60, 193–207.
- Hagarová, I.; Kudrík, I. Optimization of extraction procedure with nonionic surfactant for determination of trace lead in waters. Chem. Listy 2016, 110, 504–510.
- Hagarová, I. Utilization of Supramolecular Solvents in the Extraction of Metals. Chem. Listy 2014, 108, 949–955.
- Zhang, T.; Liu, J.-M.; Huang, X.-F.; Xia, B.; Su, C.-Y.; Luo, G.-F.; Xu, Y.-W.; Wu, Y.-X.; Mao, Z.-W.; Qiu, R.-L. Chelant extraction of heavy metals from contaminated soils using new selective EDTA derivatives. J. Hazard. Mater. 2013, 262, 464–471.
- Hiller, E.; Jurkovič, Ľ.; Faragó, T.; Vítková, M.; Tóth, R.; Komárek, M. Contaminated soils of different natural pH and industrial origin: The role of (nano) iron- and manganese-based amendments in As, Sb, Pb, and Zn leachability. Environ. Pollut. 2021, 285, 117268.
- Chmielewska, E.; Tylus, W.; Bujdoš, M. Study of Mono- and Bimetallic Fe and Mn Oxide-Supported Clinoptilolite for Improved Pb(II) Removal. Molecules 2021, 26, 4143.
- Dudová, J.; Bujdoš, M. Study of selenium sorption on iron oxide hydroxides. Chem. Listy 2015, 109, 770–774.
- Hagarová, I.; Nemček, L. Application of Metallic Nanoparticles and Their Hybrids as Innovative Sorbents for Separation and Pre-concentration of Trace Elements by Dispersive Micro-Solid Phase Extraction: A Minireview. Front. Chem. 2021, 9, 672755.
- Pamukcu, S. In Situ Soil and Sediment Remediation. In Handbook of Environmental Engineering; John Wiley & Sons: Hoboken, NJ, USA, 2018; pp. 209–248.
- Song, T.-S.; Zhang, J.; Hou, S.; Wang, H.; Zhang, D.; Li, S.; Xie, J. In situ electrokinetic remediation of toxic metal-contaminated soil driven by solid phase microbial fuel cells with a wheat straw addition. J. Appl. Chem. Biotechnol. 2018, 93, 2860–2867.
- You, Y.; Dou, J.; Xue, Y.; Jin, N.; Yang, K. Chelating Agents in Assisting Phytoremediation of Uranium-Contaminated Soils: A Review. Sustainability 2022, 14, 6379.
- Zhou, W.; Shen, B.; Meng, F.; Liu, S.; Zhang, Y. Coagulation enhancement of exopolysaccharide secreted by an Antarctic sea-ice bacterium on dye wastewater. Sep. Purif. Technol. 2010, 76, 215–221.
- Kumar, A.S.; Mody, K.; Jha, B. Bacterial exopolysaccharides—A perception. J. Basic Microbiol. 2007, 47, 103–117.
- Li, C.; Yu, Y.; Fang, A.; Feng, D.; Du, M.; Tang, A.; Chen, S.; Li, A. Insight into biosorption of heavy metals by extracellular polymer substances and the improvement of the efficacy: A review. Lett. Appl. Microbiol. 2021.
- Hagarová, I. Utilization of biosurfactants in remediation of environmental media contaminated with heavy metals. Chem. Listy 2015, 109, 431–436.
- Villela, H.D.M.; Peixoto, R.S.; Soriano, A.U.; Carmo, F.L. Microbial bioremediation of oil contaminated seawater: A survey of patent deposits and the characterization of the top genera applied. Sci. Total Environ. 2019, 666, 743–758.
- Al Disi, Z.; Al-Ghouti, M.A.; Zouari, N. Investigating the simultaneous removal of hydrocarbons and heavy metals by highly adapted Bacillus and Pseudomonas strains. Environ. Technol. Innov. 2022, 27, 102513.
- Braud, A.; Jézéquel, K.; Bazot, S.; Lebeau, T. Enhanced phytoextraction of an agricultural Cr- and Pb-contaminated soil by bioaugmentation with siderophore-producing bacteria. Chemosphere 2009, 74, 280–286.
- Chlebek, D.; Płociniczak, T.; Gobetti, S.; Kumor, A.; Hupert-Kocurek, K.; Pacwa-Płociniczak, M. Analysis of the genome of the heavy metal resistant and hydrocarbon-degrading rhizospheric Pseudomonas qingdaonensis zcr6 strain and assessment of its plant-growth-promoting traits. Int. J. Mol. Sci. 2022, 23, 214.
- Moore, E.R.B.; Tindall, B.J.; Martins Dos Santos, V.A.P.; Pieper, D.H.; Ramos, J.-L.; Palleroni, N.J. Nonmedical: Pseudomonas. In The Prokaryotes: A Handbook on the Biology of Bacteria Volume 6: Proteobacteria: Gamma Subclass; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 646–703.
- Abdelbary, S.; Elgamal, M.S.; Farrag, A. Trends in Heavy Metals Tolerance and Uptake by Pseudomonas aeruginosa. In Pseudomonas aeruginosa—An Armory Within; Sriramulu, D., Ed.; IntechOpen: London, UK, 2018.
- Nouha, K.; Kumar, R.S.; Balasubramanian, S.; Tyagi, R.D. Critical review of EPS production, synthesis and composition for sludge flocculation. J. Environ. Sci. 2018, 66, 225–245.
- Hay, I.D.; Rehman, Z.U.; Moradali, M.F.; Wang, Y.; Rehm, B.H.A. Microbial alginate production, modification and its applications. Microb. Biotechnol. 2013, 6, 637–650.
- Wu, Q.; Tun, H.M.; Leung, F.C.-C.; Shah, N.P. Genomic insights into high exopolysaccharide-producing dairy starter bacterium Streptococcus thermophilus ASCC 1275. Sci. Rep. 2014, 4, 4974.
- Osemwegie, O.O.; Adetunji, C.O.; Ayeni, E.A.; Adejobi, O.I.; Arise, R.O.; Nwonuma, C.O.; Oghenekaro, A.O. Exopolysaccharides from bacteria and fungi: Current status and perspectives in Africa. Heliyon 2020, 6, e04205.
- Rehm, B.H.A. Bacterial polymers: Biosynthesis, modifications and applications. Nat. Rev. Microbiol. 2010, 8, 578–592.
- Ohman, D.E. Molecular genetics of exopolysaccharide production by mucoid Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. 1986, 5, 6–10.
- Kasak, P.; Sasová, J.; Shoheeduzzaman, R.; Baig, M.T.; Alyafei, A.A.H.A.; Tkac, J. Influence of direct electric field on PMCG-alginate-based microcapsule. Emergent Mater. 2021, 4, 769–779.
- Kasak, P.; Danko, M.; Zavahir, S.; Mrlik, M.; Xiong, Y.; Yousaf, A.B.; Lai, W.-F.; Krupa, I.; Tkac, J.; Rogach, A.L. Identification of molecular fluorophore as a component of carbon dots able to induce gelation in a fluorescent multivalent-metal-ion-free alginate hydrogel. Sci. Rep. 2019, 9, 15080.
- Franklin, M.; Nivens, D.; Weadge, J.; Howell, P. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2011, 2, 167.
- May, T.B.; Shinabarger, D.; Boyd, A.; Chakrabarty, A.M. Identification of amino acid residues involved in the activity of phosphomannose isomerase-guanosine 5′-diphospho-D-mannose pyrophosphorylase. A bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J. Biol. Chem. 1994, 269, 4872–4877.
- Zielinski, N.A.; Chakrabarty, A.M.; Berry, A. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 1991, 266, 9754–9763.
- Tavares, I.M.; Leitão, J.H.; Fialho, A.M.; Sá-Correia, I. Pattern of changes in the activity of enzymes of GDP-D-mannuronic acid synthesis and in the level of transcription of algA, algC and algD genes accompanying the loss and emergence of mucoidy in Pseudomonas aeruginosa. Res. Microbiol. 1999, 150, 105–116.
- Oglesby, L.L.; Jain, S.; Ohman, D.E. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology 2008, 154, 1605–1615.
- Remminghorst, U.; Rehm, B.H.A. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 2006, 580, 3883–3888.
- Maharaj, R.; May, T.B.; Shang-Kwei, W.; Chakrabarty, A.M. Sequence of the alg8 and alg44 genes involved in the synthesis of alginate by Pseudomonas aeruginosa. Gene 1993, 136, 267–269.
- Hay, I.D.; Remminghorst, U.; Rehm, B.H.A. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009, 75, 1110–1120.
- Nivens, D.E.; Ohman, D.E.; Williams, J.; Franklin, M.J. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 2001, 183, 1047–1057.
- Franklin, M.J.; Chitnis, C.E.; Gacesa, P.; Sonesson, A.; White, D.C.; Ohman, D.E. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J. Bacteriol. 1994, 176, 1821–1830.
- Robles-Price, A.; Wong Thiang, Y.; Sletta, H.; Valla, S.; Schiller Neal, L. AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 2004, 186, 7369–7377.
- Schiller, N.L.; Monday, S.R.; Boyd, C.M.; Keen, N.T.; Ohman, D.E. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): Cloning, sequencing, and expression in Escherichia coli. J. Bacteriol. 1993, 175, 4780–4789.
- Whitney, J.C.; Hay, I.D.; Li, C.; Eckford, P.D.W.; Robinson, H.; Amaya, M.F.; Wood, L.F.; Ohman, D.E.; Bear, C.E.; Rehm, B.H.; et al. Structural basis for alginate secretion across the bacterial outer membrane. Proc. Natl. Acad. Sci. USA 2011, 108, 13083–13088.
- Keiski, C.-L.; Harwich, M.; Jain, S.; Neculai, A.M.; Yip, P.; Robinson, H.; Whitney, J.C.; Riley, L.; Burrows, L.L.; Ohman, D.E.; et al. AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure 2010, 18, 265–273.
- Myszka, K.; Czaczyk, K. Characterization of adhesive exopolysaccharide (EPS) produced by Pseudomonas aeruginosa under starvation conditions. Curr. Microbiol. 2009, 58, 541–546.
- Grobe, S.; Wingender, J.; Flemming, H.-C. Capability of mucoid Pseudomonas aeruginosa to survive in chlorinated water. Int. J. Hyg. Environ. Health 2001, 204, 139–142.
- Grobe, S.; Wingender, J.; Trüper, H.G. Characterization of mucoid Pseudomonas aeruginosa strains isolated from technical water systems. J. Appl. Bacteriol. 1995, 79, 94–102.
- Wozniak, D.J.; Wyckoff, T.J.O.; Starkey, M.; Keyser, R.; Azadi, P.; O’Toole, G.A.; Parsek, M.R. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 2003, 100, 7907–7912.
- Colvin, K.M.; Irie, Y.; Tart, C.S.; Urbano, R.; Whitney, J.C.; Ryder, C.; Howell, P.L.; Wozniak, D.J.; Parsek, M.R. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2012, 14, 1913–1928.
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358.
- Le Mauff, F.; Razvi, E.; Reichhardt, C.; Sivarajah, P.; Parsek, M.R.; Howell, P.L.; Sheppard, D.C. The Pel polysaccharide is predominantly composed of a dimeric repeat of α-1,4 linked galactosamine and N-acetylgalactosamine. Commun. Biol. 2022, 5, 502.
- Friedman, L.; Kolter, R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 2004, 51, 675–690.
- Ghafoor, A.; Jordens, Z.; Rehma, H.A.B. Role of pelf in pel polysaccharide biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2013, 79, 2968–2978.
- Whitfield Gregory, B.; Marmont Lindsey, S.; Ostaszewski, A.; Rich Jacquelyn, D.; Whitney John, C.; Parsek Matthew, R.; Harrison Joe, J.; Howell, P.L. Pel Polysaccharide Biosynthesis Requires an Inner Membrane Complex Comprised of PelD, PelE, PelF, and PelG. J. Bacteriol. 2020, 202, e00684-19.
- Low, K.E.; Howell, P.L. Gram-negative synthase-dependent exopolysaccharide biosynthetic machines. Curr. Opin. Struct. Biol. 2018, 53, 32–44.
- Colvin Kelly, M.; Alnabelseya, N.; Baker, P.; Whitney John, C.; Howell, P.L.; Parsek Matthew, R. PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2013, 195, 2329–2339.
- Marmont, L.S.; Rich, J.D.; Whitney, J.C.; Whitfield, G.B.; Almblad, H.; Robinson, H.; Parsek, M.R.; Harrison, J.J.; Howell, P.L. Oligomeric lipoprotein PelC guides Pel polysaccharide export across the outer membrane of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, 2892–2897.
- Byrd, M.S.; Sadovskaya, I.; Vinogradov, E.; Lu, H.; Sprinkle, A.B.; Richardson, S.H.; Ma, L.; Ralston, B.; Parsek, M.R.; Anderson, E.M.; et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 2009, 73, 622–638.
- Lee, H.-J.; Chang, H.-Y.; Venkatesan, N.; Peng, H.-L. Identification of amino acid residues important for the phosphomannose isomerase activity of PslB in Pseudomonas aeruginosa PAO1. FEBS Lett. 2008, 582, 3479–3483.
- Baker, P.; Whitfield, G.B.; Hill, P.J.; Little, D.J.; Pestrak, M.J.; Robinson, H.; Wozniak, D.J.; Howell, P.L. Characterization of the Pseudomonas aeruginosa glycoside hydrolase PslG reveals that Its levels are critical for Psl polysaccharide biosynthesis and biofilm formation. J. Biol. Chem. 2015, 290, 28374–28387.
- Wu, H.; Wang, D.; Tang, M.; Ma, L.Z. The advance of assembly of exopolysaccharide Psl biosynthesis machinery in Pseudomonas aeruginosa. MicrobiologyOpen 2019, 8, e857.
More
Information
Subjects:
Environmental Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
832
Revisions:
2 times
(View History)
Update Date:
21 Jun 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




