
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Iasonas Dermitzakis | -- | 3483 | 2023-05-16 18:07:04 | | | |
| 2 | Peter Tang | -1 word(s) | 3482 | 2023-05-17 05:58:47 | | |
Video Upload Options
Being immune privileged, the central nervous system (CNS) is populated by unique parenchymal and non-parenchymal tissue-resident macrophages, namely, microglia and border-associated macrophages (BAMs), respectively. BAMs are found in the choroid plexus, meningeal and perivascular spaces, playing critical roles in maintaining CNS homeostasis while being phenotypically and functionally distinct from microglial cells. Although the ontogeny of microglia has been largely determined, BAMs need comparable scrutiny as they have been discovered and have not been thoroughly explored. Shedding light on the molecular cues and drivers orchestrating BAM generation is essential for delineating their cellular identity. BAMs are receiving more attention since they are gradually incorporated into neurodegenerative and neuroinflammatory disease evaluations. Understanding the ontogeny of BAMs and their involvement in CNS diseases paves the way for targeted therapeutic strategies and precision medicine.
1. Introduction
2. Origin of BAMs during Embryogenesis and Adulthood
3. Molecular Drives Orchestrating BAM Development
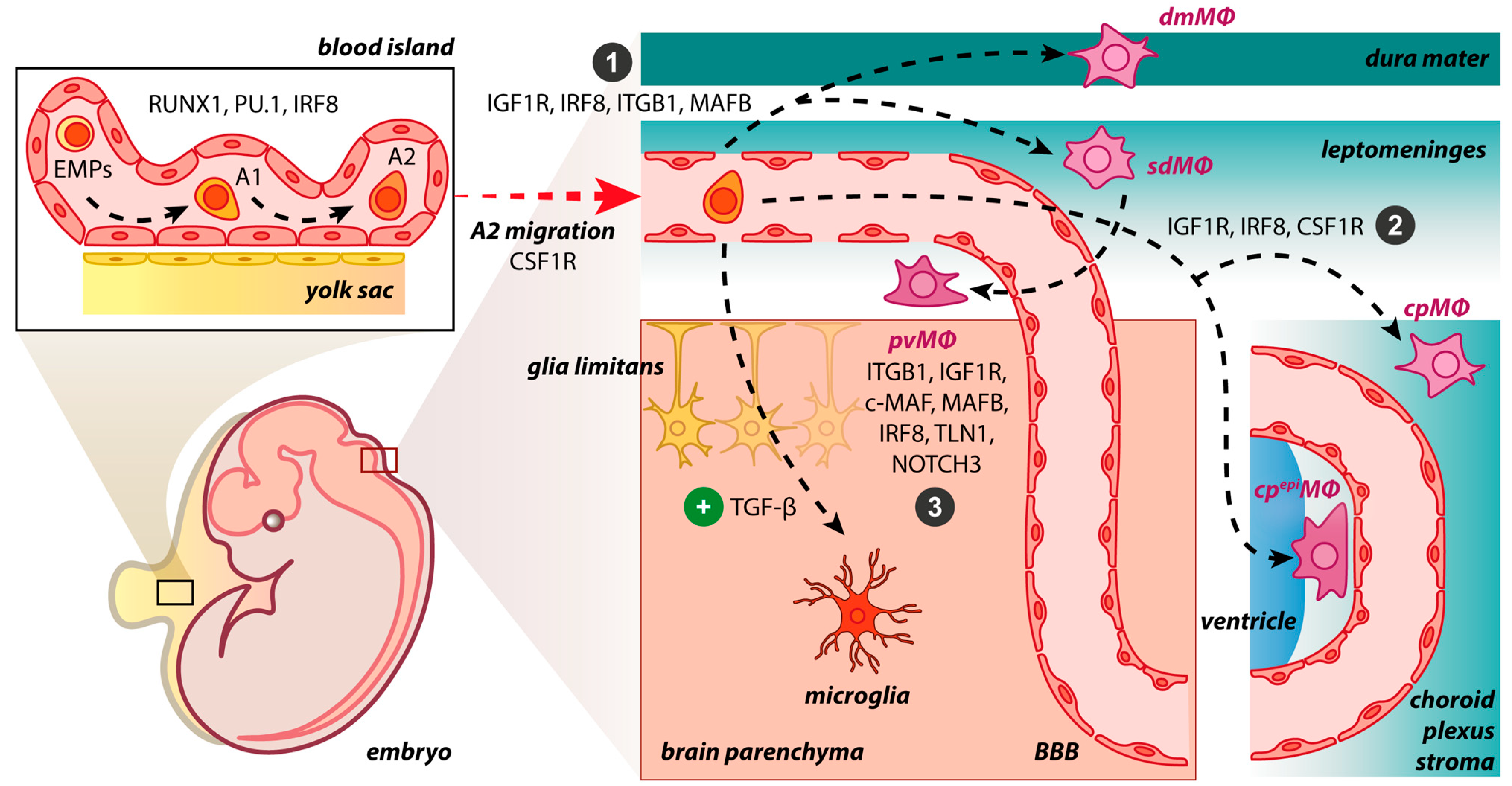
4. BAMs vs. Microglia
|
Cell Type |
Morphology |
Motility |
Cell-Specific Markers |
|---|---|---|---|
|
Microglia |
Ramified in homeostasis; Amoeboid in inflammation |
Cell bodies with limited-motility but highly dynamic processes in homeostasis; Highly phagocytic in inflammation |
SIGLEC-H+, P2RY12+, HEXB+, TMEM119+, ANXA3+, SALL1+ |
|
pvΜΦ |
Slightly elongated cell bodies |
Non-motile cell bodies with extending and retracting projections through the blood vessel wall in homeostasis; Dendritic-like processes in inflammation |
CD206+, CD38+, LYVE1+, CD36+, CD163+, CD169+ |
|
dmΜΦ |
Elongated; Spindle-shaped cells; Few thick membrane projections; Dendriform |
Limited motility and highly dynamic protrusions in homeostasis; Extending projections in inflammation |
|
|
sdΜΦ |
Elongated; Amoeboid; Spindle-shaped cells; Few thick membrane projections |
Limited motility and highly dynamic protrusions in homeostasis; Extending projections in inflammation |
|
|
cpΜΦ |
Star-like shape |
Unknown |
|
|
cpepiΜΦ |
Round; Bipolar; Stellate |
Unknown |
5. BAMs in Neurological Diseases and Promising Therapies
The implication of BAMs in the pathogenesis of CNS diseases, especially in neurodegeneration and neuroinflammation, is a rapidly emerging field of research. Although the precise role of BAMs in diseases is not yet elucidated, recent studies have addressed their potential involvement in several pathological conditions such as Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, and stroke. Further experimental studies are needed to delineate the exact pathophysiological mechanism through which methodical manipulation of BAMs can halt or even reverse the progression of the aforementioned debilitating CNS diseases.
5.1. BAMs in Alzheimer’s Disease
5.2. BAMs in Parkinson’s Disease
5.3. BAMs in Multiple Sclerosis
5.4. BAMs in Other CNS Diseases
References
- Mastorakos, P.; McGavern, D. The Anatomy and Immunology of Vasculature in the Central Nervous System. Sci. Immunol. 2019, 4, eaav0492.
- Li, Q.; Barres, B.A. Microglia and Macrophages in Brain Homeostasis and Disease. Nat. Rev. Immunol. 2018, 18, 225–242.
- Galloway, D.A.; Phillips, A.E.M.; Owen, D.R.J.; Moore, C.S. Phagocytosis in the Brain: Homeostasis and Disease. Front. Immunol. 2019, 10, 790.
- Casano, A.M.; Peri, F. Microglia: Multitasking Specialists of the Brain. Dev. Cell 2015, 32, 469–477.
- Mrdjen, D.; Pavlovic, A.; Hartmann, F.J.; Schreiner, B.; Utz, S.G.; Leung, B.P.; Lelios, I.; Heppner, F.L.; Kipnis, J.; Merkler, D.; et al. High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity 2018, 48, 380–395.e6.
- Bechmann, I.; Kwidzinski, E.; Kovac, A.D.; Simbürger, E.; Horvath, T.; Gimsa, U.; Dirnagl, U.; Priller, J.; Nitsch, R. Turnover of Rat Brain Perivascular Cells. Exp. Neurol. 2001, 168, 242–249.
- Mato, M.; Ookawara, S.; Kurihara, K. Uptake of Exogenous Substances and Marked Infoldings of the Fluorescent Granular Pericyte in Cerebral Fine Vessels. Am. J. Anat. 1980, 157, 329–332.
- Goldmann, T.; Wieghofer, P.; Jordão, M.J.C.; Prutek, F.; Hagemeyer, N.; Frenzel, K.; Amann, L.; Staszewski, O.; Kierdorf, K.; Krueger, M.; et al. Origin, Fate and Dynamics of Macrophages at Central Nervous System Interfaces. Nat. Immunol. 2016, 17, 797–805.
- McKinsey, G.L.; Lizama, C.O.; Keown-Lang, A.E.; Niu, A.; Santander, N.; Larpthaveesarp, A.; Chee, E.; Gonzalez, F.F.; Arnold, T.D. A New Genetic Strategy for Targeting Microglia in Development and Disease. eLife 2020, 9, e54590.
- Masuda, T.; Amann, L.; Sankowski, R.; Staszewski, O.; Lenz, M.; d’Errico, P.; Snaidero, N.; Costa Jordão, M.J.; Böttcher, C.; Kierdorf, K.; et al. Novel Hexb-Based Tools for Studying Microglia in the CNS. Nat. Immunol. 2020, 21, 802–815.
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Gokhan, S.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; et al. Fate Mapping Analysis Reveals That Adult Microglia Derive from Primitive Macrophages. Science 2010, 330, 841–845.
- Jordão, M.J.C.; Sankowski, R.; Brendecke, S.M.; Locatelli, G.; Tai, Y.-H.; Tay, T.L.; Schramm, E.; Armbruster, S.; Hagemeyer, N.; Groß, O.; et al. Single-Cell Profiling Identifies Myeloid Cell Subsets with Distinct Fates during Neuroinflammation. Science 2019, 363, eaat7554.
- Yona, S.; Kim, K.-W.; Wolf, Y.; Mildner, A.; Varol, D.; Breker, M.; Strauss-Ayali, D.; Viukov, S.; Guilliams, M.; Misharin, A.; et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity 2013, 38, 79–91.
- Van Hove, H.; Martens, L.; Scheyltjens, I.; De Vlaminck, K.; Pombo Antunes, A.R.; De Prijck, S.; Vandamme, N.; De Schepper, S.; Van Isterdael, G.; Scott, C.L.; et al. A Single-Cell Atlas of Mouse Brain Macrophages Reveals Unique Transcriptional Identities Shaped by Ontogeny and Tissue Environment. Nat. Neurosci. 2019, 22, 1021–1035.
- Zelco, A.; Börjesson, V.; de Kanter, J.K.; Lebrero-Fernandez, C.; Lauschke, V.M.; Rocha-Ferreira, E.; Nilsson, G.; Nair, S.; Svedin, P.; Bemark, M.; et al. Single-Cell Atlas Reveals Meningeal Leukocyte Heterogeneity in the Developing Mouse Brain. Genes Dev. 2021, 35, 1190–1207.
- Mildenberger, W.; Stifter, S.A.; Greter, M. Diversity and Function of Brain-Associated Macrophages. Curr. Opin. Immunol. 2022, 76, 102181.
- van Furth, R.; Cohn, Z.A.; Hirsch, J.G.; Humphrey, J.H.; Spector, W.G.; Langevoort, H.L. The Mononuclear Phagocyte System: A New Classification of Macrophages, Monocytes, and Their Precursor Cells. Bull. World Health Organ. 1972, 46, 845–852.
- Hoeffel, G.; Chen, J.; Lavin, Y.; Low, D.; Almeida, F.F.; See, P.; Beaudin, A.E.; Lum, J.; Low, I.; Forsberg, E.C.; et al. C-Myb+ Erythro-Myeloid Progenitor-Derived Fetal Monocytes Give Rise to Adult Tissue-Resident Macrophages. Immunity 2015, 42, 665–678.
- Hickey, W.F.; Kimura, H. Perivascular Microglial Cells of the CNS Are Bone Marrow-Derived and Present Antigen in Vivo. Science 1988, 239, 290–292.
- Utz, S.G.; See, P.; Mildenberger, W.; Thion, M.S.; Silvin, A.; Lutz, M.; Ingelfinger, F.; Rayan, N.A.; Lelios, I.; Buttgereit, A.; et al. Early Fate Defines Microglia and Non-Parenchymal Brain Macrophage Development. Cell 2020, 181, 557–573.e18.
- Ginhoux, F.; Guilliams, M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 2016, 44, 439–449.
- Shemer, A.; Grozovski, J.; Tay, T.L.; Tao, J.; Volaski, A.; Süß, P.; Ardura-Fabregat, A.; Gross-Vered, M.; Kim, J.-S.; David, E.; et al. Engrafted Parenchymal Brain Macrophages Differ from Microglia in Transcriptome, Chromatin Landscape and Response to Challenge. Nat. Commun. 2018, 9, 5206.
- Stremmel, C.; Schuchert, R.; Wagner, F.; Thaler, R.; Weinberger, T.; Pick, R.; Mass, E.; Ishikawa-Ankerhold, H.C.; Margraf, A.; Hutter, S.; et al. Yolk Sac Macrophage Progenitors Traffic to the Embryo during Defined Stages of Development. Nat. Commun. 2018, 9, 75.
- Dermitzakis, I.; Manthou, M.E.; Meditskou, S.; Tremblay, M.-È.; Petratos, S.; Zoupi, L.; Boziki, M.; Kesidou, E.; Simeonidou, C.; Theotokis, P. Origin and Emergence of Microglia in the CNS—An Interesting (Hi)Story of an Eccentric Cell. Curr. Issues Mol. Biol. 2023, 45, 2609–2628.
- Shao, F.; Wang, X.; Wu, H.; Wu, Q.; Zhang, J. Microglia and Neuroinflammation: Crucial Pathological Mechanisms in Traumatic Brain Injury-Induced Neurodegeneration. Front. Aging Neurosci. 2022, 14, 825086.
- Sims, R.; van der Lee, S.J.; Naj, A.C.; Bellenguez, C.; Badarinarayan, N.; Jakobsdottir, J.; Kunkle, B.W.; Boland, A.; Raybould, R.; Bis, J.C.; et al. Rare Coding Variants in PLCG2, ABI3, and TREM2 Implicate Microglial-Mediated Innate Immunity in Alzheimer’s Disease. Nat. Genet. 2017, 49, 1373–1384.
- Doorn, K.J.; Moors, T.; Drukarch, B.; van de Berg, W.D.; Lucassen, P.J.; van Dam, A.-M. Microglial Phenotypes and Toll-like Receptor 2 in the Substantia Nigra and Hippocampus of Incidental Lewy Body Disease Cases and Parkinson’s Disease Patients. Acta Neuropathol. Commun. 2014, 2, 90.
- Smajić, S.; Prada-Medina, C.A.; Landoulsi, Z.; Ghelfi, J.; Delcambre, S.; Dietrich, C.; Jarazo, J.; Henck, J.; Balachandran, S.; Pachchek, S.; et al. Single-Cell Sequencing of Human Midbrain Reveals Glial Activation and a Parkinson-Specific Neuronal State. Brain 2022, 145, 964–978.
- Prineas, J.W.; Kwon, E.E.; Cho, E.-S.; Sharer, L.R.; Barnett, M.H.; Oleszak, E.L.; Hoffman, B.; Morgan, B.P. Immunopathology of Secondary-Progressive Multiple Sclerosis. Ann. Neurol. 2001, 50, 646–657.
- Zrzavy, T.; Hametner, S.; Wimmer, I.; Butovsky, O.; Weiner, H.L.; Lassmann, H. Loss of ‘Homeostatic’ Microglia and Patterns of Their Activation in Active Multiple Sclerosis. Brain 2017, 140, 1900–1913.
- Prinz, M.; Masuda, T.; Wheeler, M.A.; Quintana, F.J. Microglia and Central Nervous System-Associated Macrophages-From Origin to Disease Modulation. Annu. Rev. Immunol. 2021, 39, 251–277.
- Bartholomäus, I.; Kawakami, N.; Odoardi, F.; Schläger, C.; Miljkovic, D.; Ellwart, J.W.; Klinkert, W.E.F.; Flügel-Koch, C.; Issekutz, T.B.; Wekerle, H.; et al. Effector T Cell Interactions with Meningeal Vascular Structures in Nascent Autoimmune CNS Lesions. Nature 2009, 462, 94–98.
- Wasser, B.; Luchtman, D.; Löffel, J.; Robohm, K.; Birkner, K.; Stroh, A.; Vogelaar, C.F.; Zipp, F.; Bittner, S. CNS-Localized Myeloid Cells Capture Living Invading T Cells during Neuroinflammation. J. Exp. Med. 2020, 217, e20190812.
- De Schepper, S.; Ge, J.Z.; Crowley, G.; Ferreira, L.S.S.; Garceau, D.; Toomey, C.E.; Sokolova, D.; Rueda-Carrasco, J.; Shin, S.-H.; Kim, J.-S.; et al. Perivascular Cells Induce Microglial Phagocytic States and Synaptic Engulfment via SPP1 in Mouse Models of Alzheimer’s Disease. Nat. Neurosci. 2023, 26, 406–415.
- Prinz, M.; Schmidt, H.; Mildner, A.; Knobeloch, K.-P.; Hanisch, U.-K.; Raasch, J.; Merkler, D.; Detje, C.; Gutcher, I.; Mages, J.; et al. Distinct and Nonredundant In Vivo Functions of IFNAR on Myeloid Cells Limit Autoimmunity in the Central Nervous System. Immunity 2008, 28, 675–686.
- Rajan, W.D.; Wojtas, B.; Gielniewski, B.; Miró-Mur, F.; Pedragosa, J.; Zawadzka, M.; Pilanc, P.; Planas, A.M.; Kaminska, B. Defining Molecular Identity and Fates of CNS-Border Associated Macrophages after Ischemic Stroke in Rodents and Humans. Neurobiol. Dis. 2020, 137, 104722.
- Bechmann, I.; Priller, J.; Kovac, A.; Böntert, M.; Wehner, T.; Klett, F.F.; Bohsung, J.; Stuschke, M.; Dirnagl, U.; Nitsch, R. Immune Surveillance of Mouse Brain Perivascular Spaces by Blood-Borne Macrophages. Eur. J. Neurosci. 2001, 14, 1651–1658.
- Hickey, W.F.; Vass, K.; Lassmann, H. Bone Marrow-Derived Elements in the Central Nervous System: An Immunohistochemical and Ultrastructural Survey of Rat Chimeras. J. Neuropathol. Exp. Neurol. 1992, 51, 246–256.
- Mass, E.; Ballesteros, I.; Farlik, M.; Halbritter, F.; Günther, P.; Crozet, L.; Jacome-Galarza, C.E.; Händler, K.; Klughammer, J.; Kobayashi, Y.; et al. Specification of Tissue-Resident Macrophages during Organogenesis. Science 2016, 353, aaf4238.
- Masuda, T.; Amann, L.; Monaco, G.; Sankowski, R.; Staszewski, O.; Krueger, M.; Del Gaudio, F.; He, L.; Paterson, N.; Nent, E.; et al. Specification of CNS Macrophage Subsets Occurs Postnatally in Defined Niches. Nature 2022, 604, 740–748.
- Cugurra, A.; Mamuladze, T.; Rustenhoven, J.; Dykstra, T.; Beroshvili, G.; Greenberg, Z.J.; Baker, W.; Papadopoulos, Z.; Drieu, A.; Blackburn, S.; et al. Skull and Vertebral Bone Marrow Are Myeloid Cell Reservoirs for the Meninges and CNS Parenchyma. Science 2021, 373, eabf7844.
- Wu, X.; Saito, T.; Saido, T.C.; Barron, A.M.; Ruedl, C. Microglia and CD206+ Border-Associated Mouse Macrophages Maintain Their Embryonic Origin during Alzheimer’s Disease. eLife 2021, 10, e71879.
- Huang, G.; Zhang, P.; Hirai, H.; Elf, S.; Yan, X.; Chen, Z.; Koschmieder, S.; Okuno, Y.; Dayaram, T.; Growney, J.D.; et al. PU.1 Is a Major Downstream Target of AML1 (RUNX1) in Adult Mouse Hematopoiesis. Nat. Genet. 2008, 40, 51–60.
- Kierdorf, K.; Erny, D.; Goldmann, T.; Sander, V.; Schulz, C.; Perdiguero, E.G.; Wieghofer, P.; Heinrich, A.; Riemke, P.; Hölscher, C.; et al. Microglia Emerge from Erythromyeloid Precursors via Pu.1- and Irf8-Dependent Pathways. Nat. Neurosci. 2013, 16, 273–280.
- Herbomel, P.; Thisse, B.; Thisse, C. Zebrafish Early Macrophages Colonize Cephalic Mesenchyme and Developing Brain, Retina, and Epidermis through a M-CSF Receptor-Dependent Invasive Process. Dev. Biol. 2001, 238, 274–288.
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a Unique TGF-β–Dependent Molecular and Functional Signature in Microglia. Nat. Neurosci. 2014, 17, 131–143.
- Ivan, D.C.; Berve, K.C.; Walthert, S.; Monaco, G.; Borst, K.; Bouillet, E.; Ferreira, F.; Lee, H.; Steudler, J.; Buch, T.; et al. Insulin-like Growth Factor-1 Receptor Controls the Function of CNS-Resident Macrophages and Their Contribution to Neuroinflammation. Acta Neuropathol. Commun. 2023, 11, 35.
- Hamilton, J.A.; Achuthan, A. Colony Stimulating Factors and Myeloid Cell Biology in Health and Disease. Trends Immunol. 2013, 34, 81–89.
- Hamilton, J.A. Colony-Stimulating Factors in Inflammation and Autoimmunity. Nat. Rev. Immunol. 2008, 8, 533–544.
- Sauter, K.A.; Bouhlel, M.A.; O’Neal, J.; Sester, D.P.; Tagoh, H.; Ingram, R.M.; Pridans, C.; Bonifer, C.; Hume, D.A. The Function of the Conserved Regulatory Element within the Second Intron of the Mammalian Csf1r Locus. PLoS ONE 2013, 8, e54935.
- Munro, D.A.D.; Bradford, B.M.; Mariani, S.A.; Hampton, D.W.; Vink, C.S.; Chandran, S.; Hume, D.A.; Pridans, C.; Priller, J. CNS Macrophages Differentially Rely on an Intronic Csf1r Enhancer for Their Development. Dev. Camb. Engl. 2020, 147, dev194449.
- Rojo, R.; Raper, A.; Ozdemir, D.D.; Lefevre, L.; Grabert, K.; Wollscheid-Lengeling, E.; Bradford, B.; Caruso, M.; Gazova, I.; Sánchez, A.; et al. Deletion of a Csf1r Enhancer Selectively Impacts CSF1R Expression and Development of Tissue Macrophage Populations. Nat. Commun. 2019, 10, 3215.
- Hagemeyer, N.; Kierdorf, K.; Frenzel, K.; Xue, J.; Ringelhan, M.; Abdullah, Z.; Godin, I.; Wieghofer, P.; Costa Jordão, M.J.; Ulas, T.; et al. Transcriptome-Based Profiling of Yolk Sac-Derived Macrophages Reveals a Role for Irf8 in Macrophage Maturation. EMBO J. 2016, 35, 1730–1744.
- Moura Silva, H.; Kitoko, J.Z.; Queiroz, C.P.; Kroehling, L.; Matheis, F.; Yang, K.L.; Reis, B.S.; Ren-Fielding, C.; Littman, D.R.; Bozza, M.T.; et al. C-MAF-Dependent Perivascular Macrophages Regulate Diet-Induced Metabolic Syndrome. Sci. Immunol. 2021, 6, eabg7506.
- Wang, Q.; Zhao, N.; Kennard, S.; Lilly, B. Notch2 and Notch3 Function Together to Regulate Vascular Smooth Muscle Development. PloS ONE 2012, 7, e37365.
- Dermitzakis, I.; Manthou, M.E.; Meditskou, S.; Miliaras, D.; Kesidou, E.; Boziki, M.; Petratos, S.; Grigoriadis, N.; Theotokis, P. Developmental Cues and Molecular Drivers in Myelinogenesis: Revisiting Early Life to Re-Evaluate the Integrity of CNS Myelin. Curr. Issues Mol. Biol. 2022, 44, 3208–3237.
- Colonna, M.; Butovsky, O. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468.
- Tremblay, M.-È.; Stevens, B.; Sierra, A.; Wake, H.; Bessis, A.; Nimmerjahn, A. The Role of Microglia in the Healthy Brain. J. Neurosci. 2011, 31, 16064–16069.
- Butovsky, O.; Weiner, H.L. Microglial Signatures and Their Role in Health and Disease. Nat. Rev. Neurosci. 2018, 19, 622–635.
- Lenz, K.M.; Nelson, L.H. Microglia and Beyond: Innate Immune Cells As Regulators of Brain Development and Behavioral Function. Front. Immunol. 2018, 9, 698.
- Chen, Z.; Trapp, B.D. Microglia and Neuroprotection. J. Neurochem. 2016, 136 (Suppl. 1), 10–17.
- Goddery, E.N.; Fain, C.E.; Lipovsky, C.G.; Ayasoufi, K.; Yokanovich, L.T.; Malo, C.S.; Khadka, R.H.; Tritz, Z.P.; Jin, F.; Hansen, M.J.; et al. Microglia and Perivascular Macrophages Act as Antigen Presenting Cells to Promote CD8 T Cell Infiltration of the Brain. Front. Immunol. 2021, 12, 726421.
- Fabriek, B.O.; Van Haastert, E.S.; Galea, I.; Polfliet, M.M.J.; Döpp, E.D.; Van Den Heuvel, M.M.; Van Den Berg, T.K.; De Groot, C.J.A.; Van Der Valk, P.; Dijkstra, C.D. CD163-Positive Perivascular Macrophages in the Human CNS Express Molecules for Antigen Recognition and Presentation. Glia 2005, 51, 297–305.
- Mato, M.; Ookawara, S.; Sakamoto, A.; Aikawa, E.; Ogawa, T.; Mitsuhashi, U.; Masuzawa, T.; Suzuki, H.; Honda, M.; Yazaki, Y.; et al. Involvement of Specific Macrophage-Lineage Cells Surrounding Arterioles in Barrier and Scavenger Function in Brain Cortex. Proc. Natl. Acad. Sci. USA 1996, 93, 3269–3274.
- Lim, H.Y.; Lim, S.Y.; Tan, C.K.; Thiam, C.H.; Goh, C.C.; Carbajo, D.; Chew, S.H.S.; See, P.; Chakarov, S.; Wang, X.N.; et al. Hyaluronan Receptor LYVE-1-Expressing Macrophages Maintain Arterial Tone through Hyaluronan-Mediated Regulation of Smooth Muscle Cell Collagen. Immunity 2018, 49, 326–341.e7.
- He, H.; Mack, J.J.; Güç, E.; Warren, C.M.; Squadrito, M.L.; Kilarski, W.W.; Baer, C.; Freshman, R.D.; McDonald, A.I.; Ziyad, S.; et al. Perivascular Macrophages Limit Permeability. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 2203–2212.
- Galanternik, M.V.; Castranova, D.; Gore, A.V.; Blewett, N.H.; Jung, H.M.; Stratman, A.N.; Kirby, M.R.; Iben, J.; Miller, M.F.; Kawakami, K.; et al. A Novel Perivascular Cell Population in the Zebrafish Brain. eLife 2017, 6, e24369.
- Liu, C.; Wu, C.; Yang, Q.; Gao, J.; Li, L.; Yang, D.; Luo, L. Macrophages Mediate the Repair of Brain Vascular Rupture through Direct Physical Adhesion and Mechanical Traction. Immunity 2016, 44, 1162–1176.
- Jais, A.; Solas, M.; Backes, H.; Chaurasia, B.; Kleinridders, A.; Theurich, S.; Mauer, J.; Steculorum, S.M.; Hampel, B.; Goldau, J.; et al. Myeloid-Cell-Derived VEGF Maintains Brain Glucose Uptake and Limits Cognitive Impairment in Obesity. Cell 2016, 165, 882–895.
- Mato, M.; Ookawara, S.; Sano, M.; Fukuda, S. Uptake of Fat by Fluorescent Granular Perithelial Cells in Cerebral Cortex after Administration of Fat Rich Chow. Experientia 1982, 38, 1496–1498.
- Cummings, J. Alzheimer’s Disease Diagnostic Criteria: Practical Applications. Alzheimers Res. Ther. 2012, 4, 35.
- Tiwari, S.; Atluri, V.; Kaushik, A.; Yndart, A.; Nair, M. Alzheimer’s Disease: Pathogenesis, Diagnostics, and Therapeutics. Int. J. Nanomed. 2019, 14, 5541–5554.
- Muralidar, S.; Ambi, S.V.; Sekaran, S.; Thirumalai, D.; Palaniappan, B. Role of Tau Protein in Alzheimer’s Disease: The Prime Pathological Player. Int. J. Biol. Macromol. 2020, 163, 1599–1617.
- Weber, S.A.; Patel, R.K.; Lutsep, H.L. Cerebral Amyloid Angiopathy: Diagnosis and Potential Therapies. Expert Rev. Neurother. 2018, 18, 503–513.
- Hawkes, C.A.; McLaurin, J. Selective Targeting of Perivascular Macrophages for Clearance of Beta-Amyloid in Cerebral Amyloid Angiopathy. Proc. Natl. Acad. Sci. USA 2009, 106, 1261–1266.
- Park, L.; Uekawa, K.; Garcia-Bonilla, L.; Koizumi, K.; Murphy, M.; Pistik, R.; Younkin, L.; Younkin, S.; Zhou, P.; Carlson, G.; et al. Brain Perivascular Macrophages Initiate the Neurovascular Dysfunction of Alzheimer Aβ Peptides. Circ. Res. 2017, 121, 258–269.
- Simon, D.K.; Tanner, C.M.; Brundin, P. Parkinson Disease Epidemiology, Pathology, Genetics, and Pathophysiology. Clin. Geriatr. Med. 2020, 36, 1–12.
- Lee, J.-W.; Chun, W.; Lee, H.J.; Kim, S.-M.; Min, J.-H.; Kim, D.-Y.; Kim, M.-O.; Ryu, H.W.; Lee, S.U. The Role of Microglia in the Development of Neurodegenerative Diseases. Biomedicines 2021, 9, 1449.
- Gómez-Benito, M.; Granado, N.; García-Sanz, P.; Michel, A.; Dumoulin, M.; Moratalla, R. Modeling Parkinson’s Disease with the Alpha-Synuclein Protein. Front. Pharmacol. 2020, 11, 356.
- Guo, M.; Wang, J.; Zhao, Y.; Feng, Y.; Han, S.; Dong, Q.; Cui, M.; Tieu, K. Microglial Exosomes Facilitate α-Synuclein Transmission in Parkinson’s Disease. Brain J. Neurol. 2020, 143, 1476–1497.
- Schonhoff, A.; Figge, D.; Williams, G.; Jurkuvenaite, A.; Gallups, N.; Childers, G.; Webster, J.; Standaert, D.; Goldman, J.; Harms, A. Border-Associated Macrophages Mediate the Neuroinflammatory Response in an Alpha-Synuclein Model of Parkinson Disease. bioRxiv 2022.
- Qin, H.; Buckley, J.A.; Li, X.; Liu, Y.; Fox, T.H.; Meares, G.P.; Yu, H.; Yan, Z.; Harms, A.S.; Li, Y.; et al. Inhibition of the JAK/STAT Pathway Protects Against α-Synuclein-Induced Neuroinflammation and Dopaminergic Neurodegeneration. J. Neurosci. 2016, 36, 5144–5159.
- Walton, C.; King, R.; Rechtman, L.; Kaye, W.; Leray, E.; Marrie, R.A.; Robertson, N.; La Rocca, N.; Uitdehaag, B.; van der Mei, I.; et al. Rising Prevalence of Multiple Sclerosis Worldwide: Insights from the Atlas of MS, Third Edition. Mult. Scler. J. 2020, 26, 1816–1821.
- Mirmosayyeb, O.; Brand, S.; Barzegar, M.; Afshari-Safavi, A.; Nehzat, N.; Shaygannejad, V.; Sadeghi Bahmani, D. Clinical Characteristics and Disability Progression of Early- and Late-Onset Multiple Sclerosis Compared to Adult-Onset Multiple Sclerosis. J. Clin. Med. 2020, 9, 1326.
- Polman, C.H.; Reingold, S.C.; Banwell, B.; Clanet, M.; Cohen, J.A.; Filippi, M.; Fujihara, K.; Havrdova, E.; Hutchinson, M.; Kappos, L.; et al. Diagnostic Criteria for Multiple Sclerosis: 2010 Revisions to the McDonald Criteria. Ann. Neurol. 2011, 69, 292–302.
- Alcina, A.; Fedetz, M.; Vidal-Cobo, I.; Andrés-León, E.; García-Sánchez, M.-I.; Barroso-del-Jesus, A.; Eichau, S.; Gil-Varea, E.; Villar, L.-M.; Saiz, A.; et al. Identification of the Genetic Mechanism That Associates L3MBTL3 to Multiple Sclerosis. Hum. Mol. Genet. 2022, 31, 2155–2163.
- Jandric, D.; Lipp, I.; Paling, D.; Rog, D.; Castellazzi, G.; Haroon, H.; Parkes, L.; Parker, G.J.M.; Tomassini, V.; Muhlert, N. Mechanisms of Network Changes in Cognitive Impairment in Multiple Sclerosis. Neurology 2021, 97, e1886–e1897.
- Correale, J.; Marrodan, M.; Ysrraelit, M.C. Mechanisms of Neurodegeneration and Axonal Dysfunction in Progressive Multiple Sclerosis. Biomedicines 2019, 7, 14.
- Jäckle, K.; Zeis, T.; Schaeren-Wiemers, N.; Junker, A.; van der Meer, F.; Kramann, N.; Stadelmann, C.; Brück, W. Molecular Signature of Slowly Expanding Lesions in Progressive Multiple Sclerosis. Brain 2020, 143, 2073–2088.
- Kamma, E.; Lasisi, W.; Libner, C.; Ng, H.S.; Plemel, J.R. Central Nervous System Macrophages in Progressive Multiple Sclerosis: Relationship to Neurodegeneration and Therapeutics. J. Neuroinflamm. 2022, 19, 45.
- Miedema, A.; Gerrits, E.; Brouwer, N.; Jiang, Q.; Kracht, L.; Meijer, M.; Nutma, E.; Peferoen-Baert, R.; Pijnacker, A.T.E.; Wesseling, E.M.; et al. Brain Macrophages Acquire Distinct Transcriptomes in Multiple Sclerosis Lesions and Normal Appearing White Matter. Acta Neuropathol. Commun. 2022, 10, 8.
- Locatelli, G.; Theodorou, D.; Kendirli, A.; Jordão, M.J.C.; Staszewski, O.; Phulphagar, K.; Cantuti-Castelvetri, L.; Dagkalis, A.; Bessis, A.; Simons, M.; et al. Mononuclear Phagocytes Locally Specify and Adapt Their Phenotype in a Multiple Sclerosis Model. Nat. Neurosci. 2018, 21, 1196–1208.
- Theotokis, P.; Lourbopoulos, A.; Touloumi, O.; Lagoudaki, R.; Kofidou, E.; Nousiopoulou, E.; Poulatsidou, K.-N.; Kesidou, E.; Tascos, N.; Spandou, E.; et al. Time Course and Spatial Profile of Nogo-A Expression in Experimental Autoimmune Encephalomyelitis in C57BL/6 Mice. J. Neuropathol. Exp. Neurol. 2012, 71, 907–920.
- Theotokis, P.; Touloumi, O.; Lagoudaki, R.; Nousiopoulou, E.; Kesidou, E.; Siafis, S.; Tselios, T.; Lourbopoulos, A.; Karacostas, D.; Grigoriadis, N.; et al. Nogo Receptor Complex Expression Dynamics in the Inflammatory Foci of Central Nervous System Experimental Autoimmune Demyelination. J. Neuroinflamm. 2016, 13, 265.
- Montilla, A.; Zabala, A.; Er-Lukowiak, M.; Rissiek, B.; Magnus, T.; Rodriguez-Iglesias, N.; Sierra, A.; Matute, C.; Domercq, M. Microglia and Meningeal Macrophages Depletion Delays the Onset of Experimental Autoimmune Encephalomyelitis. Cell Death Dis. 2023, 14, 16.
- McCarthy, D.P.; Richards, M.H.; Miller, S.D. Mouse Models of Multiple Sclerosis: Experimental Autoimmune Encephalomyelitis and Theiler’s Virus-Induced Demyelinating Disease. Methods Mol. Biol. 2012, 900, 381–401.
- Donninelli, G.; Saraf-Sinik, I.; Mazziotti, V.; Capone, A.; Grasso, M.G.; Battistini, L.; Reynolds, R.; Magliozzi, R.; Volpe, E. Interleukin-9 Regulates Macrophage Activation in the Progressive Multiple Sclerosis Brain. J. Neuroinflamm. 2020, 17, 149.
- Roczkowsky, A.; Doan, M.A.L.; Hlavay, B.; Mamik, M.K.; Branton, W.G.; McKenzie, B.A.; Saito, L.B.; Schmitt, L.; Eitzen, G.; Di Cara, F.; et al. Peroxisome Injury in Multiple Sclerosis: Protective Effects of 4-Phenylbutyrate in CNS-Associated Macrophages. J. Neurosci. Off. J. Soc. Neurosci. 2022, 42, 7152–7165.
- Grajchen, E.; Hendriks, J.J.A.; Bogie, J.F.J. The Physiology of Foamy Phagocytes in Multiple Sclerosis. Acta Neuropathol. Commun. 2018, 6, 124.
- Haidar, M.; Loix, M.; Vanherle, S.; Dierckx, T.; Vangansewinkel, T.; Gervois, P.; Wolfs, E.; Lambrichts, I.; Bogie, J.F.J.; Hendriks, J.J.A. Targeting Lipophagy in Macrophages Improves Repair in Multiple Sclerosis. Autophagy 2022, 18, 2697–2710.
- Pedragosa, J.; Salas-Perdomo, A.; Gallizioli, M.; Cugota, R.; Miró-Mur, F.; Briansó, F.; Justicia, C.; Pérez-Asensio, F.; Marquez-Kisinousky, L.; Urra, X.; et al. CNS-Border Associated Macrophages Respond to Acute Ischemic Stroke Attracting Granulocytes and Promoting Vascular Leakage. Acta Neuropathol. Commun. 2018, 6, 76.
- Wan, H.; Brathwaite, S.; Ai, J.; Hynynen, K.; Macdonald, R.L. Role of Perivascular and Meningeal Macrophages in Outcome following Experimental Subarachnoid Hemorrhage. J. Cereb. Blood Flow Metab. 2021, 41, 1842–1857.
- Kolosowska, N.; Keuters, M.H.; Wojciechowski, S.; Keksa-Goldsteine, V.; Laine, M.; Malm, T.; Goldsteins, G.; Koistinaho, J.; Dhungana, H. Peripheral Administration of IL-13 Induces Anti-Inflammatory Microglial/Macrophage Responses and Provides Neuroprotection in Ischemic Stroke. Neurotherapeutics 2019, 16, 1304–1319.





