
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jaebong Kim | -- | 2261 | 2023-05-04 10:45:54 | | | |
| 2 | Lindsay Dong | Meta information modification | 2261 | 2023-05-06 07:40:38 | | |
Video Upload Options
Fibroblast growth factors (FGFs) encode a large family of growth factor proteins that activate several intracellular signaling pathways to control diverse physiological functions. The human genome encodes 22 FGFs that share a high sequence and structural homology with those of other vertebrates. FGFs orchestrate diverse biological functions by regulating cellular differentiation, proliferation, and migration. Dysregulated FGF signaling may contribute to several pathological conditions, including cancer. Notably, FGFs exhibit wide functional diversity among different vertebrates spatiotemporally. A comparative study of FGF receptor ligands and their diverse roles in vertebrates ranging from embryonic development to pathological conditions may expand our understanding of FGF.
1. Introduction
2. Gene Organization and Protein Length
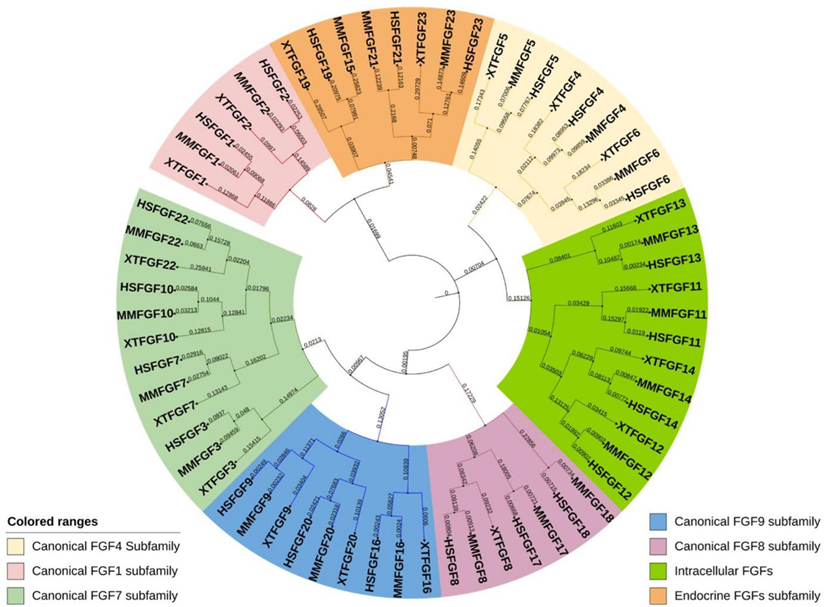
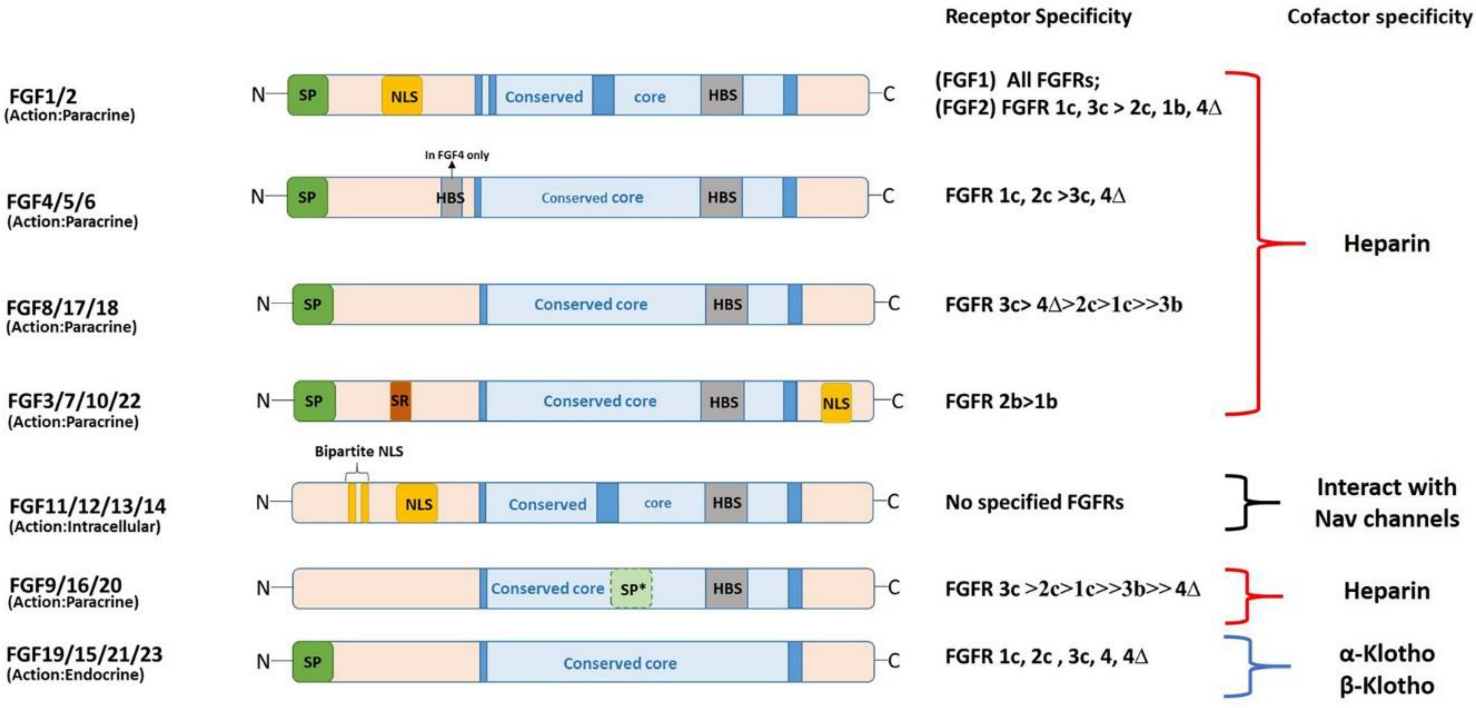
3. Structural and Functional Diversity
4. FGF Signaling in Early Development
4.1. FGF and Mesoderm Specification
4.2. FGF and Neural Specification
4.3. FGF Signaling in Metabolism and Diseases (Cancer)
4.3.1. FGF Signaling in Metabolism
4.3.2. FGF Signaling in Various Types of Cancer
References
- Itoh, N.; Ornitz, D. Functional evolutionary history of the mouseFgf gene family. Dev. Dyn. 2008, 237, 18–27.
- Johnson, D.E.; Williams, L.T. Structural and Functional Diversity in the FGf Receptor Multigene Family. Adv. Cancer Res. 1993, 60, 1–41.
- Morrison, R.S.; Sharma, A.; de Vellis, J.; Bradshaw, R.A. Basic fibroblast growth factor supports the survival of cerebral cortical neurons in primary culture. Proc. Natl. Acad. Sci. USA 1986, 83, 7537–7541.
- Folkman, J.; Klagsbrun, M. Angiogenic factors. Science 1987, 235, 442–447.
- Teven, C.M.; Farina, E.M.; Rivas, J.; Reid, R.R. Fibroblast growth factor (FGF) signaling in development and skeletal diseases. Genes Dis. 2014, 1, 199–213.
- Xie, Y.; Su, N.; Yang, J.; Tan, Q.; Huang, S.; Jin, M.; Ni, Z.; Zhang, B.; Zhang, D.; Luo, F.; et al. FGF/FGFR signaling in health and disease. Signal Transduct. Target. Ther. 2020, 5, 181.
- Ornitz, D.M.; Itoh, N. Fibroblast growth factors. Genome Biol. 2001, 2, REVIEWS3005.
- Ornitz, D.M. FGFs, heparan sulfate and FGFRs: Complex interactions essential for development. Bioessays 2000, 22, 108–112.
- Kuro-O, M. Endocrine FGFs and Klothos: Emerging concepts. Trends Endocrinol. Metab. 2008, 19, 239–245.
- Ornitz, D.M.; Itoh, N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 215–266.
- Goetz, R.; Mohammadi, M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 2013, 14, 166–180.
- Kumar, V.; Goutam, R.S.; Park, S.; Lee, U.; Kim, J. Functional Roles of FGF Signaling in Early Development of Vertebrate Embryos. Cells 2021, 10, 2148.
- Itoh, N.; Ornitz, D.M. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004, 20, 563–569.
- Kiefer, P.; Acland, P.; Pappin, D.; Peters, G.; Dickson, C. Competition between nuclear localization and secretory signals determines the subcellular fate of a single CUG-initiated form of FGF3. EMBO J. 1994, 13, 4126–4136.
- Arnaud, E.; Touriol, C.; Boutonnet, C.; Gensac, M.-C.; Vagner, S.; Prats, H.; Prats, A.-C. A New 34-Kilodalton Isoform of Human Fibroblast Growth Factor 2 Is Cap Dependently Synthesized by Using a Non-AUG Start Codon and Behaves as a Survival Factor. Mol. Cell. Biol. 1999, 19, 505–514.
- Sunmonu, N.A.; Li, K.; Li, J.Y. Numerous isoforms of Fgf8 reflect its multiple roles in the developing brain. J. Cell. Physiol. 2011, 226, 1722–1726.
- Itoh, N.; Ornitz, D.M. Fibroblast growth factors: From molecular evolution to roles in development, metabolism and disease. J. Biochem. 2011, 149, 121–130.
- Powers, C.J.; McLeskey, S.W.; Wellstein, A. Fibroblast growth factors, their receptors and signaling. Endocr.-Relat. Cancer 2000, 7, 165–197.
- Plotnikov, A.N.; Hubbard, S.R.; Schlessinger, J.; Mohammadi, M. Crystal Structures of Two FGF-FGFR Complexes Reveal the Determinants of Ligand-Receptor Specificity. Cell 2000, 101, 413–424.
- Mohammadi, M.; Olsen, S.K.; Ibrahimi, O.A. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005, 16, 107–137.
- Laestander, C.; Engström, W. Role of fibroblast growth factors in elicitation of cell responses. Cell Prolif. 2014, 47, 3–11.
- Kolli, V.; Paul, S.; Sarkar, N. An Overview on Fibroblast Growth Factors: Structural, Functional and Therapeutic Implications. Curr. Proteom. 2015, 12, 144–151.
- Lea, R.; Papalopulu, N.; Amaya, E.; Dorey, K. Temporal and spatial expression of FGF ligands and receptors during Xenopus development. Dev. Dyn. 2009, 238, 1467–1479.
- Yayon, A.; Klagsbrun, M.; Esko, J.D.; Leder, P.; Ornitz, D.M. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 1991, 64, 841–848.
- Mohammadi, M.; Olsen, S.K.; Goetz, R. A protein canyon in the FGF–FGF receptor dimer selects from an à la carte menu of heparan sulfate motifs. Curr. Opin. Struct. Biol. 2005, 15, 506–516.
- Sacco, A.; Federico, C.; Giacomini, A.; Caprio, C.; Maccarinelli, F.; Todoerti, K.; Favasuli, V.; Anastasia, A.; Motta, M.; Russo, D.; et al. Halting the FGF/FGFR axis leads to antitumor activity in Waldenström macroglobulinemia by silencing MYD88. Blood 2021, 137, 2495–2508.
- Planque, N. Nuclear trafficking of secreted factors and cell-surface receptors: New pathways to regulate cell proliferation and differentiation, and involvement in cancers. Cell Commun. Signal. 2006, 4, 7.
- Olsnes, S.; Klingenberg, O.; Wie˛dłocha, A. Transport of Exogenous Growth Factors and Cytokines to the Cytosol and to the Nucleus. Physiol. Rev. 2003, 83, 163–182.
- Rodriguez-Enfedaque, A.; Bouleau, S.; Laurent, M.; Courtois, Y.; Mignotte, B.; Vayssière, J.-L.; Renaud, F. FGF1 nuclear translocation is required for both its neurotrophic activity and its p53-dependent apoptosis protection. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2009, 1793, 1719–1727.
- Bouleau, S.; Grimal, H.; Rincheval, V.; Godefroy, N.; Mignotte, B.; Vayssière, J.-L.; Renaud, F. FGF1 inhibits p53-dependent apoptosis and cell cycle arrest via an intracrine pathway. Oncogene 2005, 24, 7839–7849.
- Oulion, S.; Bertrand, S.; Escriva, H. Evolution of the FGF Gene Family. Int. J. Evol. Biol. 2012, 2012, 298147.
- Zhang, X.; Ibrahimi, O.A.; Olsen, S.; Umemori, H.; Mohammadi, M.; Ornitz, D.M. Receptor Specificity of the Fibroblast Growth Factor Family. J. Biol. Chem. 2006, 281, 15694–15700.
- Ornitz, D.M.; Xu, J.; Colvin, J.S.; McEwen, D.G.; MacArthur, C.A.; Coulier, F.; Gao, G.; Goldfarb, M. Receptor Specificity of the Fibroblast Growth Factor Family. J. Biol. Chem. 1996, 271, 15292–15297.
- Amaya, E.; Musci, T.J.; Kirschner, M.W. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in xenopus embryos. Cell 1991, 66, 257–270.
- Amaya, E.; A Stein, P.; Musci, T.J.; Kirschner, M.W. FGF signalling in the early specification of mesoderm in Xenopus. Development 1993, 118, 477–487.
- Fletcher, R.B.; Harland, R.M. The role of FGF signaling in the establishment and maintenance of mesodermal gene expression in Xenopus. Dev. Dyn. 2008, 237, 1243–1254.
- Fan, T.-P.; Ting, H.-C.; Yu, J.-K.; Su, Y.-H. Reiterative use of FGF signaling in mesoderm development during embryogenesis and metamorphosis in the hemichordate Ptychodera flava. BMC Evol. Biol. 2018, 18, 120.
- Henrique, D.; Abranches, E.; Verrier, L.; Storey, K.G. Neuromesodermal progenitors and the making of the spinal cord. Development 2015, 142, 2864–2875.
- Wilson, V.I. Olivera-Martinez, and K.G. Storey, Stem cells, signals and vertebrate body axis extension. Development 2009, 136, 1591–1604.
- del Corral, R.D.; Morales, A.V. The Multiple Roles of FGF Signaling in the Developing Spinal Cord. Front. Cell Dev. Biol. 2017, 5, 58.
- Delfino-Machín, M.; Lunn, J.S.; Breitkreuz, D.N.; Akai, J.; Storey, K. Specification and maintenance of the spinal cord stem zone. Development 2005, 132, 4273–4283.
- Karabagli, H.; Karabagli, P.; Ladher, R.K.; Schoenwolf, G.C. Comparison of the expression patterns of several fibroblast growth factors during chick gastrulation and neurulation. Anat. Embryol. 2002, 205, 365–370.
- Bertrand, V.; Hudson, C.; Caillol, D.; Popovici, C.; Lemaire, P. Neural Tissue in Ascidian Embryos Is Induced by FGF9/16/20, Acting via a Combination of Maternal GATA and Ets Transcription Factors. Cell 2003, 115, 615–627.
- Miya, T.; Nishida, H. An Ets transcription factor, HrEts, is target of FGF signaling and involved in induction of notochord, mesenchyme, and brain in ascidian embryos. Dev. Biol. 2003, 261, 25–38.
- Spemann, H.; Mangold, H. über Induktion von Embryonalanlagen durch Implantation artfremder Organisatoren. Dev. Genes Evol. 1924, 100, 599–638.
- Wilson, S.I.; Edlund, T. Neural induction: Toward a unifying mechanism. Nat. Neurosci. 2001, 4, 1161–1168.
- Stern, C.D. Neural induction: Old problem, new findings, yet more questions. Development 2005, 132, 2007–2021.
- Kliewer, S.A.; Mangelsdorf, D.J. Bile Acids as Hormones: The FXR-FGF15/19 Pathway. Dig. Dis. 2015, 33, 327–331.
- Tomlinson, E.; Fu, L.; John, L.; Hultgren, B.; Huang, X.; Renz, M.; Stephan, J.P.; Tsai, S.P.; Powell-Braxton, L.; French, D.; et al. Transgenic mice expressing human fibroblast growth factor-19 display increased metabolic rate and decreased adiposity. Endocrinology 2002, 143, 1741–1747.
- Bhatnagar, S.; Damron, H.A.; Hillgartner, F. Fibroblast Growth Factor-19, a Novel Factor That Inhibits Hepatic Fatty Acid Synthesis. J. Biol. Chem. 2009, 284, 10023–10033.
- Tacer, K.F.; Bookout, A.L.; Ding, X.; Kurosu, H.; John, G.B.; Wang, L.; Goetz, R.; Mohammadi, M.; Kuro-O, M.; Mangelsdorf, D.J.; et al. Research Resource: Comprehensive Expression Atlas of the Fibroblast Growth Factor System in Adult Mouse. Mol. Endocrinol. 2010, 24, 2050–2064.
- Coskun, T.; Bina, H.A.; Schneider, M.A.; Dunbar, J.D.; Hu, C.C.; Chen, Y.; Moller, D.E.; Kharitonenkov, A. Fibroblast Growth Factor 21 Corrects Obesity in Mice. Endocrinology 2008, 149, 6018–6027.
- Xu, J.; Lloyd, D.J.; Hale, C.; Stanislaus, S.; Chen, M.; Sivits, G.; Vonderfecht, S.; Hecht, R.; Li, Y.-S.; Lindberg, R.A.; et al. Fibroblast Growth Factor 21 Reverses Hepatic Steatosis, Increases Energy Expenditure, and Improves Insulin Sensitivity in Diet-Induced Obese Mice. Diabetes 2009, 58, 250–259.
- Kharitonenkov, A.; Beals, J.M.; Micanovic, R.; Strifler, B.A.; Rathnachalam, R.; Wroblewski, V.J.; Li, S.; Koester, A.; Ford, A.M.; Coskun, T.; et al. Rational Design of a Fibroblast Growth Factor 21-Based Clinical Candidate, LY2405319. PLoS ONE 2013, 8, e58575.
- Shapter, A.E. The overpass syndrome. Can. Med. Assoc. J. 1992, 146, 113.
- Quarles, L.D. Skeletal secretion of FGF-23 regulates phosphate and vitamin D metabolism. Nat. Rev. Endocrinol. 2012, 8, 276–286.
- Consortium, A. Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat. Genet. 2000, 26, 345–348.
- Shimada, T.; Urakawa, I.; Yamazaki, Y.; Hasegawa, H.; Hino, R.; Yoneya, T.; Takeuchi, Y.; Fujita, T.; Fukumoto, S.; Yamashita, T. FGF-23 transgenic mice demonstrate hypophosphatemic rickets with reduced expression of sodium phosphate cotransporter type IIa. Biochem. Biophys. Res. Commun. 2004, 314, 409–414.
- Larsson, T.; Marsell, R.; Schipani, E.; Ohlsson, C.; Ljunggren, O.; Tenenhouse, H.S.; Jppner, H.; Jonsson, K.B. Transgenic mice expressing fibroblast growth factor 23 under the control of the alpha1(I) collagen promoter exhibit growth retardation, osteomalacia, and disturbed phosphate homeostasis. Endocrinology 2004, 145, 3087–3094.
- Bai, X.; Miao, D.; Li, J.; Goltzman, D.; Karaplis, A.C. Transgenic Mice Overexpressing Human Fibroblast Growth Factor 23 (R176Q) Delineate a Putative Role for Parathyroid Hormone in Renal Phosphate Wasting Disorders. Endocrinology 2004, 145, 5269–5279.
- Szlachcic, A.; Sochacka, M.; Czyrek, A.; Opalinski, L.; Krowarsch, D.; Otlewski, J.; Zakrzewska, M. Low Stability of Integrin-Binding Deficient Mutant of FGF1 Restricts Its Biological Activity. Cells 2019, 8, 899.
- Slattery, M.L.; John, E.M.; Stern, M.C.; Herrick, J.; Lundgreen, A.; Giuliano, A.R.; Hines, L.; Baumgartner, K.B.; Torres-Mejia, G.; Wolff, R.K.; et al. Associations with growth factor genes (FGF1, FGF2, PDGFB, FGFR2, NRG2, EGF, ERBB2) with breast cancer risk and survival: The Breast Cancer Health Disparities Study. Breast Cancer Res. Treat. 2013, 140, 587–601.
- Ribatti, D.; Vacca, A.; Rusnati, M.; Presta, M. The discovery of basic fibroblast growth factor/fibroblast growth factor-2 and its role in haematological malignancies. Cytokine Growth Factor Rev. 2007, 18, 327–334.
- Giulianelli, S.; Riggio, M.; Guillardoy, T.; Pérez Piñero, C.; Gorostiaga, M.A.; Sequeira, G.; Pataccini, G.; Abascal, M.F.; Toledo, M.F.; Jacobsen, B.M.; et al. FGF2 induces breast cancer growth through ligand-independent activation and recruitment of ERalpha and PRBDelta4 isoform to MYC regulatory sequences. Int. J. Cancer 2019, 145, 1874–1888.
- Hamamoto, J.; Yasuda, H.; Nonaka, Y.; Fujiwara, M.; Nakamura, Y.; Soejima, K.; Betsuyaku, T. The FGF2 aptamer inhibits the growth of FGF2-FGFR pathway driven lung cancer cells. Biochem. Biophys. Res. Commun. 2018, 503, 1330–1334.
- Wang, Y.; Becker, D. Antisense targeting of basic fibroblast growth factor and dibroblast growth factor receptor-1 in human melanomas blocks intratumoral angiogenesis and tumor growth. Nat. Med. 1997, 3, 887–893.
- Maerz, W.J.; Baselga, J.; Reuter, V.E.; Mellado, B.; Myers, M.L.; Bosl, G.J.; Spinella, M.J.; Dmitrovsky, E. FGF4 dissociates anti-tumorigenic from differentiation signals of retinoic acid in human embryonal carcinomas. Oncogene 1998, 17, 761–767.
- Yasuda, K.; Torigoe, T.; Mariya, T.; Asano, T.; Kuroda, T.; Matsuzaki, J.; Ikeda, K.; Yamauchi, M.; Emori, M.; Asanuma, H.; et al. Fibroblasts induce expression of FGF4 in ovarian cancer stem-like cells/cancer-initiating cells and upregulate their tumor initiation capacity. Lab. Investig. 2014, 94, 1355–1369.
- Huang, Y.; Wang, H.; Yang, Y. Expression of Fibroblast Growth Factor 5 (FGF5) and Its Influence on Survival of Breast Cancer Patients. Experiment 2018, 24, 3524–3530.
- Guo, S.; Jiang, S.; Epperla, N.; Ma, Y.; Maadooliat, M.; Ye, Z.; Olson, B.; Wang, M.; Kitchner, T.; Joyce, J.; et al. A gene-based recessive diplotype exome scan discovers FGF6, a novel hepcidin-regulating iron-metabolism gene. Blood 2019, 133, 1888–1898.




