Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Shujie Yang | -- | 2514 | 2023-05-03 15:35:18 | | | |
| 2 | Camila Xu | Meta information modification | 2514 | 2023-05-04 02:56:38 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Qi, A.; Lamont, L.; Liu, E.; Murray, S.D.; Meng, X.; Yang, S. Critical Functions of PHB2. Encyclopedia. Available online: https://encyclopedia.pub/entry/43705 (accessed on 07 February 2026).
Qi A, Lamont L, Liu E, Murray SD, Meng X, Yang S. Critical Functions of PHB2. Encyclopedia. Available at: https://encyclopedia.pub/entry/43705. Accessed February 07, 2026.
Qi, Amanda, Lillie Lamont, Evelyn Liu, Sarina D. Murray, Xiangbing Meng, Shujie Yang. "Critical Functions of PHB2" Encyclopedia, https://encyclopedia.pub/entry/43705 (accessed February 07, 2026).
Qi, A., Lamont, L., Liu, E., Murray, S.D., Meng, X., & Yang, S. (2023, May 03). Critical Functions of PHB2. In Encyclopedia. https://encyclopedia.pub/entry/43705
Qi, Amanda, et al. "Critical Functions of PHB2." Encyclopedia. Web. 03 May, 2023.
Copy Citation
The prohibitin (PHB) gene was initially found to be antiproliferative and able to inhibit the initiation of DNA synthesis in rat liver in 1989. The human homologue (later to be known as PHB1) was then identified, cloned, and mapped to the human chromosome 17q21. In 1994, another member of prohibitin, PHB2, was discovered on chromosome 12p13 when two proteins were found to associate with the IgM antigen receptor of B lymphocytes.
prohibitins
PHB2
essential gene
anti-apoptosis
1. Introduction
1.1. History of Prohibitin
The prohibitin (PHB) gene was initially found to be antiproliferative and able to inhibit the initiation of DNA synthesis in rat liver in 1989 [1]. The human homologue (later to be known as PHB1) was then identified, cloned, and mapped to the human chromosome 17q21 [2][3]. In 1994, another member of prohibitin, PHB2, was discovered on chromosome 12p13 when two proteins were found to associate with the IgM antigen receptor of B lymphocytes. Otherwise known as B cell receptor-associated proteins (BAPs), amino acid sequencing of BAP32 and BAP37 identified them to be PHB1 and PHB2, respectively [4]. Another name for PHB2 denoted as REA (repressor of estrogen receptor activity) was found to inhibit estrogen receptors’ (ERs) activity [5]. DNA sequencing showed that REA had the same identity as BAP37 [5]. The history of PHB2 was summarized in Figure 1.
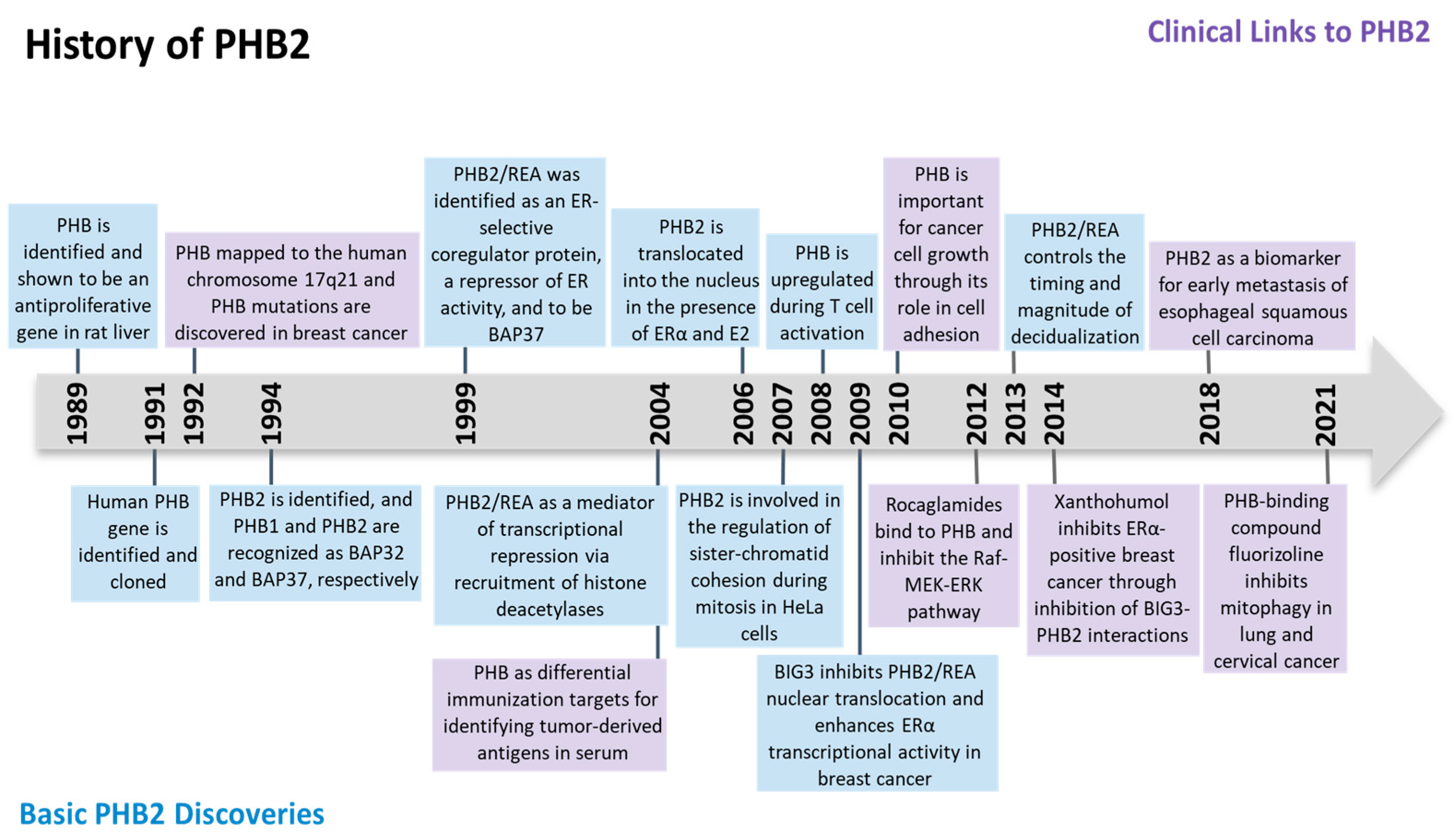
Figure 1. Timeline of major discoveries in prohibitin 2 (PHB2) studies, including basic science and clinical translational studies.
1.2. Prohibitin Family and Structure
Prohibitin (PHB) 1 and 2 belong to the Stomatin/prohibitin/flotillin/HflKC (SPFH) family, which also includes Erlin-1 and 2, podocin, stomatin, and flotillin-1 and 2. The main function of this family contributes to the formation of membrane microdomains and lipid raft-associated processes [6]. The human PHB genes encode two protein isoforms, PHB1 and PHB2, with molecular weights of 32 and 34 kDa, respectively. They both contain an evolutionarily conserved prohibitin N-terminal transmembrane domain involved in scaffolding and a C-terminal coiled-coil domain for protein–protein interactions [7]. PHB2 contains three different domains including a transmembrane domain required for mitochondrial localization, a central prohibitin domain, and an overlapping coiled-coil domain as shown in Figure 2 [3].
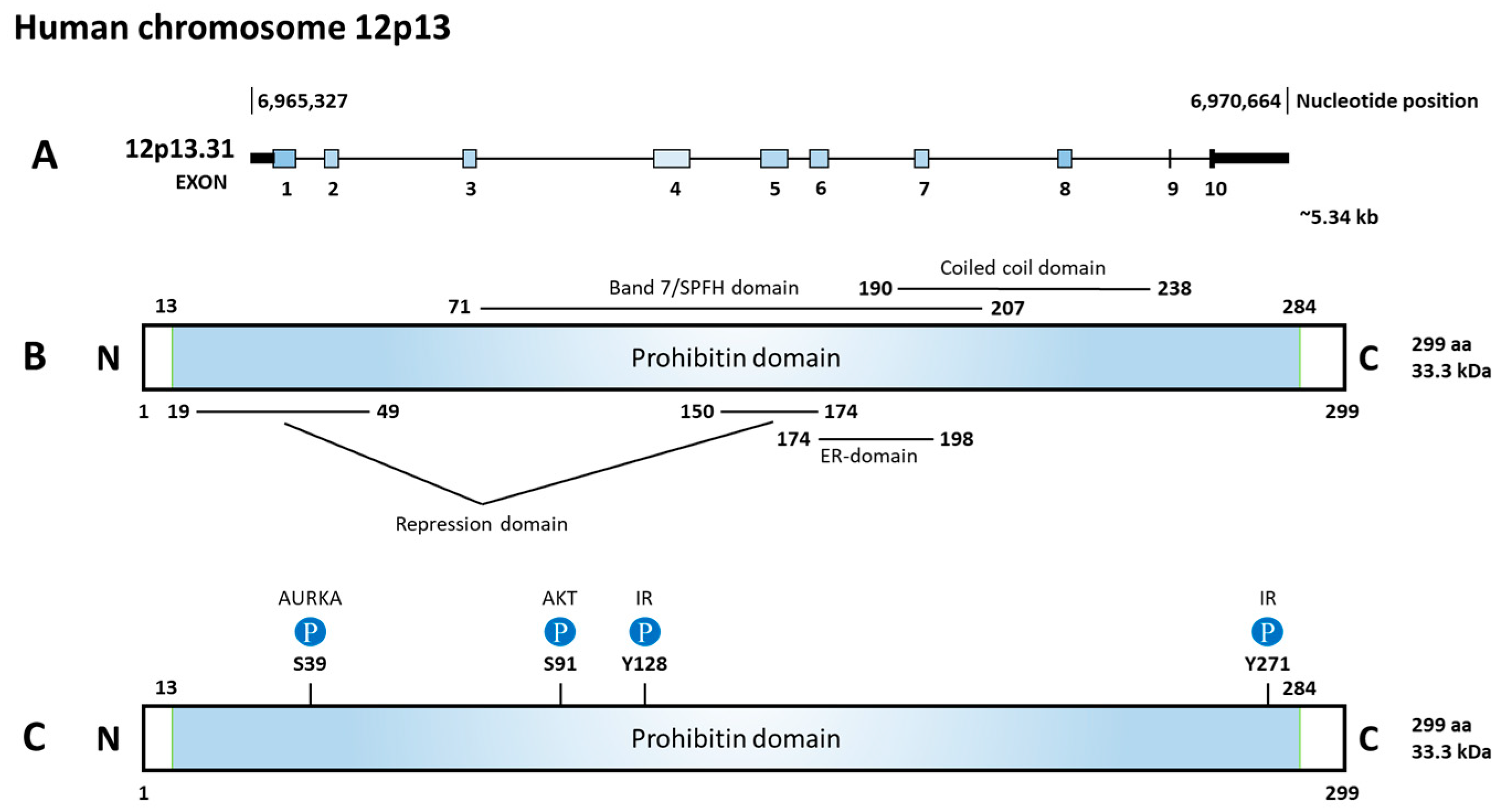
Figure 2. Diagram of gene, function domains, and phosphorylation sites of prohibitin 2 (PHB2). (A) PHB2 is located on chromosome 12p13.31, consisting of 10 exons. (B) The prohibitin domain locates from 13aa to 284aa. Inside the prohibitin domain, there are two repression domains, a Band7/SPFH domain, a coiled coil domain, and an estrogen receptor (ER)-domain. (C) Upstream regulators and the phosphorylated site on PHB2. Aurora kinase A (AURKA) phosphorylates S39, AKT serine/threonine kinase (AKT) phosphorylates S91, and insulin receptor kinase (IR) phosphorylates Y128 and Y271.
They are both expressed in the mitochondria, nucleus, and cell membrane, and are involved in the processes of cell proliferation, apoptosis, mitophagy, and metastasis. In the inner mitochondrial membrane (IMM), PHB1 and PHB2 form heterodimers assembled in ring-shaped complexes to maintain mitochondrial stability but are exported out upon signaling [8]. In the nucleus, PHB1 and PHB2 work independently to modulate transcriptional activity [9].
1.3. Prohibitin Expression
High expressions of PHB1 and PHB2 were reported in 1078 cell lines in the Cancer Cell Line Encyclopedia (CCLE) and were recognized as common essential genes in 1077 of the 1078 tested cell lines (Depmap.org/Portal). A recent publication also confirmed that PHB1 and PHB2 are common essential genes with potential mechanisms involved in protein translation initiation and transfer RNA (tRNA) ligases [10]. The average PHB2 expression is high in 1078 tested cell lines and PHB2 high expression can be derived from amplification.
In a 2018 review paper, expression of PHB1 and PHB2 was surveyed in 17 cancer types. It was reported that PHB1 and PHB2 expression levels are high in most cancer types, for both mRNA and protein [11].
1.4. PHB1
Initial studies found that PHB1 induced cell cycle arrest and inhibited cell proliferation. However, recent studies of PHB1 in esophageal squamous cell carcinoma (ESCC), gallbladder cancer, and bladder cancer exhibit promotion of cancer cell proliferation. The role of PHB1 in cancer remains inconsistent.
2. Critical Functions of PHB2
2.1. Conditional Depletion of PHB2
PHB2 is essential for cell survival. Homozygous knockout (ko) PHB2 is lethal, which demonstrates that PHB2 is indispensable for development [12][13]. Therefore, conditional ko PHB2 has been investigated in multiple model systems and has been proven to be crucial for maintaining organ function.
In pancreatic β-cells, ko PHB2 promotes mitochondrial damage and impairs insulin secretion through loss of β-cells. Thus, ko PHB2 decreases survival and induces gradual diabetes [12]. As for the heart, ko PHB2 hinders cardiac fatty acid oxidation, leading to heart failure. Conditional ko PHB2 mice had severe dilated cardiomyopathy and heart failure due to lipid droplet accumulation and mitochondrial dysfunction. Thus, PHB2 is thought to downregulate carnitine palmitoyltransferase1b (CPT1b), which is the most important enzyme of cardiac fatty acid oxidation [13]. Furthermore, forebrain specific ko PHB2 mice exhibited neurodegeneration, leading to behavioral and cognitive deficiencies. PHB2 demonstrated critical functions for the survival of neurons through mitochondrial fusion and ultrastructure [14]. Lastly, hepatocyte-specific ko PHB2 mice exhibited lipid accumulation, impaired gluconeogenesis, and liver failure. Similarly, liver PHB2-deficiency led to mitochondrial liver dysfunction [15]. Therefore, PHB2 is critical for organ development in the pancreas, heart, forebrain, and liver, because loss of PHB2 leads to organ deterioration.
2.2. Location-Dependent PHB2 Function
PHB2 is often located on the cell membrane, in the mitochondria, and in the nucleus, with distinct roles in different locations. These location-dependent functions will be described in this section and in Figure 3.
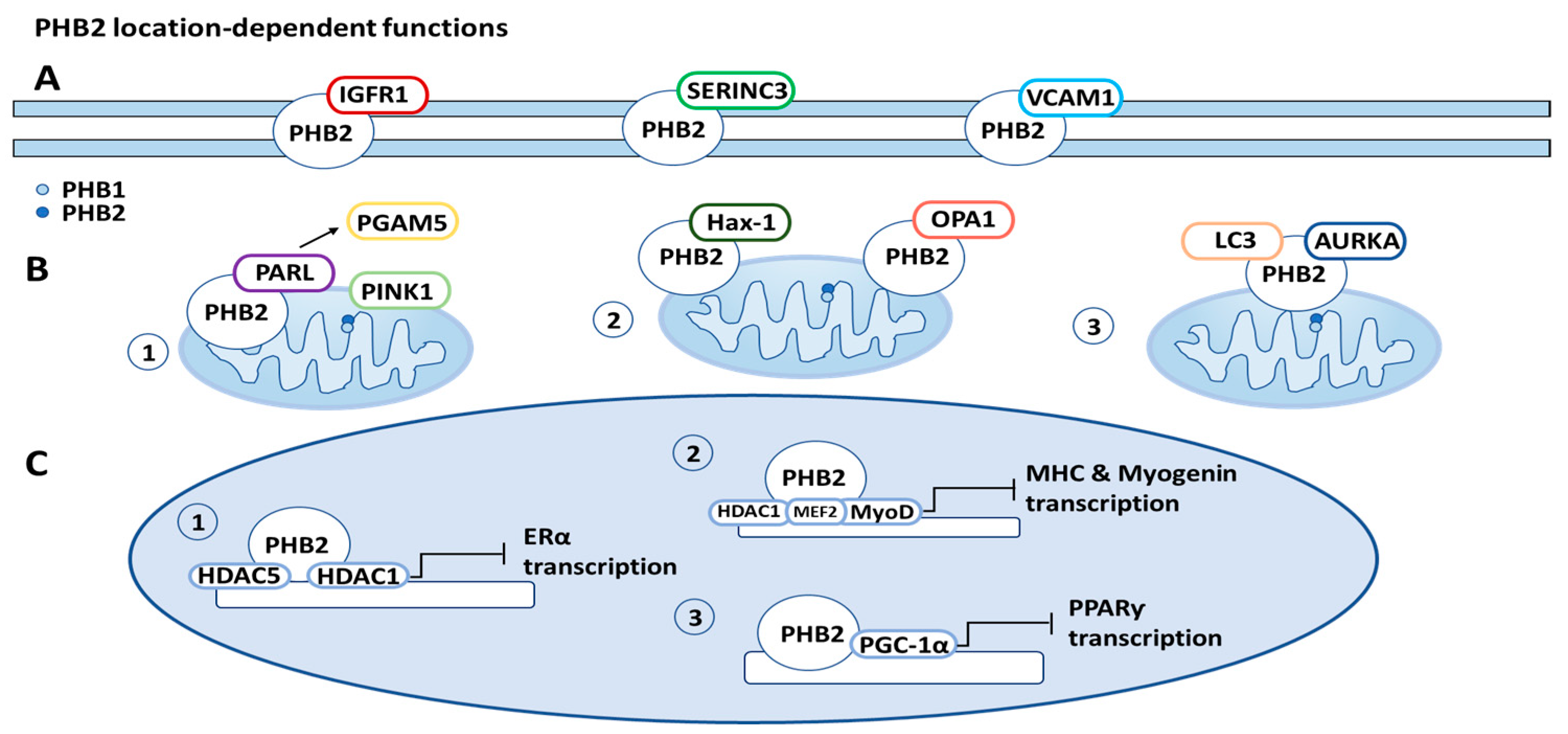
Figure 3. Prohibitin 2 (PHB2) location-related functions. (A) Cell membrane: PHB2 is associated with a variety of proteins on the cell membrane, including insulin-like growth factor-1 receptor (IGFR1), serine incorporator 3 (SERINC3), and vascular cell adhesion protein 1 (VCAM1). (B) Mitochondria: (1) PHB2 initiates mitophagy through binding presenilin-associated rhomboid-like protein (PARL), releasing PGAM5 (PGAM family member 5, mitochondrial serine/threonine protein phosphatase), and consequently stabilizing PINK1 (PTEN induced putative kinase 1). (2) PHB2 increases apoptosis resistance through binding optic atrophy 1 protein (OPA1) and HCLS1-associated protein X-1 (Hax-1). (3) PHB2 also initiates mitophagy through forming a tripartite complex with LC3 (microtubule-associated protein light chain 3) and aurora kinase A (AURKA). (C) Nucleus: (1) PHB2 recruits histone deactylases (HDACs) to the nucleus, inhibiting transcription of nuclear receptors, such as estrogen receptor-alpha (ERα). (2) PHB2 also interacts with HDAC1 (histone deacetylase 1) and forms a complex with transcription factors MEF2 (myocyte enhancing factor 2A) and MyoD (myoblast determination protein 1) to inhibit MHC (myosin heavy chain protein) and myogenin transcription. (3) PHB2 interacts with PGC-1α (peroxisome proliferator-activated receptor-ƴ coactivator), inhibiting the transcription of peroxisome proliferator-activated receptor (PPARƴ).
2.2.1. PHB2 Function on Cell Membrane
PHB2 was found to be present and expressed in multiple cellular compartments, including the plasma membrane, and was also found to be associated with specific cell membrane receptors [9][16]. PHB2 originates from an evolutionarily conserved family of proteins, including stomatins, flotillins, and the human insulin receptor [16]. Furthermore, stomatin-like proteins regulate ion channels and mechanosensation; flotillins affect signaling across the plasma membrane and regulate membrane curvature; erlins target inositol (1,4,5)-trisphosphate receptors in the ER membrane for degradation; and prohibitins and bacterial HflK/C proteins associate with AAA proteases and are involved in proteolytic processes [17]. The role of PHB2 in signal transduction at the plasma membrane was found by the association of PHB2 with the IgM receptor in B cells [17].
Another protein found to be associated with PHB2 is Serine Incorporator 3 (SERINC3), a protein primarily localized at the plasma membrane. SERINC3 was identified in complex with PHB2 by coimmunoprecipitation and Western blotting [9]. Additional cytoplasmic proteins, such as insulin-like growth factor 1 (IGFR1), and integrins, such as vascular cell adhesion protein 1 (VCAM1), were found to interact with PHB2 and are associated with integral cell membrane proteins and cellular receptors [9]. Interestingly, the identification of PHB2 and the plasma membrane were made in infectious disease studies. The PHB1:PHB2 complex was found to associate with amino acid residues 790 to 800 in the carboxyl terminus of the human immunodeficiency virus (HIV) glycoprotein [9]. These findings suggest that PHB2 can regulate nucleolus function directly as well as via transduction pathways, which may be related to the role of PHB2 in the regulation of cell proliferation [18].
2.2.2. PHB2 Function in Mitochondria
PHB2 plays a critical role in metabolically active cells, as suggested by the high expression levels in neurons, muscle, heart, and liver cells, all of which massively rely on mitochondrial function [16]. Not surprisingly, prohibitins have been discovered to be associated with countless mitochondrial mechanisms, including mitochondrial respiratory chain subunit degradation, assembly, activity of the oxidative phosphorylation system (OXPHOS), mitochondrial biogenesis, mitochondrial apoptosis, and mitophagy [7].
Mitophagy is a selective autophagy process specifically involved in the degradation of damaged or redundant mitochondria in cells to maintain cellular homeostasis [19]. Mitophagy may promote survival through the adaptation to stress by removing mitochondria that could be permeabilized to induce cell death or, conversely, it may result in cell death from the excessive elimination of mitochondria. This degradation of mitochondria has contrasting implications on tumor growth and development, making the role of mitophagy, regarding cellular fate, variable [20].
A 2017 paper published in Cell discovered that PHB2 is a IMM mitophagy receptor. It is involved in parkin-induced mitophagy by binding the autophagosomal membrane-associated protein LC3 through PHB2’s LC3-interacting region (LIR) domains. In vivo, PHB2 has an essential role in eliminating the transmission of paternally derived mitochondrial DNA to offspring in C. elegans. The mechanism through which PHB2 regulates mitophagy in paternal mitochondrial DNA is unknown. However, this confirms PHB2’s multiple roles in mitophagy, acting as a mediator of mitochondrial function as well as IMM binding to LC3 [21][22].
Yan et al. reported that in addition to functioning as a mitophagy receptor, PHB2 enhances PTEN-induced kinase 1 parkin (PINK1-PRKN)-induced mitophagy. To initiate mitophagy, PHB2 binds to PARL (presenilin-associated rhomboid-like protein), releasing PGAM5 (phosphoglycerate mutase 5) in the process, which is responsible for stabilizing PINK1 (PTEN induced putative kinase 1) on the outer mitochondrial membrane (OMM) [23].
Prohibitins also play a crucial role in inhibiting apoptosis, the process of programmed cell death. Apoptosis is essential for cells to maintain regular physiological activity [24]. Dysfunction of the mitochondria-regulated apoptosis pathway leads to cancer proliferation [25]. PHB2 interacts with optic atrophy 1 protein (OPA1), an IMM protein, which is responsible for mitochondrial fusion and cristae integrity. OPA1 influences resistance to apoptosis by regulating mitochondrial cristae remodeling [26]. Dysregulation of OPA1 processing leads to apoptosis resistance. OPA1 has two forms: long (L-OPA1) and short (S-OPA1), through proteolytic processing. Loss of L-OPA1 and accumulation of S-OPA1 results in mitochondrial fragmentation and apoptosis. OPA1 proteolytic processing is protected by PHB2 in a chaperone-like manner. It was observed that depletion of PHB2 accelerated the cleavage of L-OPA1 into S-OPA1 [15][27]. Thus, PHB2 increases resistance to apoptosis, exerting its anti-apoptotic function via stabilizing OPA1 [27].
PHB2 was also found to directly bind with Hax-1 (HCLS1-associated protein X-1), an anti-apoptotic protein, in the mitochondria. Similarly to OPA1, depletion of PHB2 leads to an increase in the degradation of Hax-1, inducing caspase-dependent apoptosis. Reduced mitochondrial Hax-1 is associated with an increased loss of mitochondrial integrity, through the activation of the caspase 9/caspase 3 pathway, resulting in apoptosis. When Hax-1 was knocked down, apoptosis occurred without causing damage to the morphology of the mitochondria. With this, it can be concluded that modified levels of PHB2 cause a decrease in mitochondrial fusion, and an increase in the formation of reactive oxygen species (ROS) and cell death [7].
Recent discoveries have brought to light the various roles that prohibitins have in cancer, many of which are linked to the elevated metabolic reliance on mitochondria respiration. Generally, cancer cells have a higher overall energy demand that is reached through an increase in cellular respiration and glycolysis. This increase leads to a cellular disproportion of ROS and Ca2+ stores. Cancer cells require maximum mitochondrial function, and in turn, their mitochondria are especially vulnerable to oxidative damage. In these circumstances, an increase in the expression of prohibitins typically results in an increase in the stability of mitochondria. The results are not always consistent. In one study, knockdown of prohibitins led to a decrease in cell division and no change in mitochondrial integrity, while another study showed that knockdown of PHB2 led to a decrease in cell proliferation as well as a major decrease in mitochondrial integrity [9].
PHB2 also forms a tripartite complex with AURKA (aurora kinase A) and LC3 (gene name is MAP1LC3, microtubule-associated proteins 1 light chain 3). The complex induces mitophagy, following phosphorylation of PHB2 on Ser39. This is referred to as AURKA-dependent mitophagy and is not correlated with the PARK2–Parkin/PINK1 pathway. Xanthohumol, a PHB2 ligand, blocks the formation of this complex by changing the distance between AURKA and LC3. Due to this, the binding of xanthohumol prevents AURKA-dependent mitochondrial loss and prevents the excessive generation of ATP that is associated with high levels of AURKA expression. This illustrates that a metabolic change occurs when AURKA is overexpressed and demonstrates the connection between mitophagy and cancer cell’s metabolic capacity [28].
Apart from OPA1, Hax-1, and AURKA, there are many other proteins that bind PHB2 in the mitochondria, including m-AAA, OMA1 proteases, and SLP-2. PHB2 also regulates numerous mitochondria functions not mentioned, such as oxidative phosphorylation, mitochondrial biogenesis, and unfolded protein response. These have been summarized and referenced in a 2019 review paper [7].
2.2.3. PHB2 Function in Nucleus
PHB1 has critical functions in the nucleus in three areas: (1) repressing transcription, (2) inducing p53 mediated transcription, and (3) mediating RAS signal transduction (through its interaction with Raf1) [29]. In comparison with PHB1, PHB2 has similar functions in the nucleus as a mediator of transcription with various transcription factors and nuclear receptors.
Within the nucleus, PHB2 plays an essential role in the regulation of transcription factors, including estrogen receptor, myocyte enhancer factor 2 (MEF2), myoblast determination protein 1 (MyoD), and peroxisome proliferator-activated receptor (PPARγ) [30][31][32]. PHB2 was discovered to inhibit COUP-TF1 and II, orphan nuclear hormone receptors (NHRs), through recruitment of histone deacetylases [33]. As COUP-TF1 and II are both pertinent to the development of embryos and the immune system, aberrant expression of PHB2 would have catastrophic repercussions regarding development [33].
Prohibitins, PHB1 and PHB2, are also significantly linked to histone deacetylases (HDACs). To repress transcription of nuclear receptors, PHB2 associates with HDAC1 and HDAC5 [33]. PHB2 recruits HDACs to the nucleus, further inhibiting the transcription of nuclear receptors. Due to this, HDAC inhibitors can prevent transcriptional inhibition, which is typically regulated by PHB2. Therefore, PHB2 can inhibit transcription through both its immediate interaction with nuclear receptors and through the recruitment of HDACs to the transcription site.
PHB2 was also found to interact with two common muscle regulation factors, MyoD and MEF2 [32]. PHB2 represses their transcriptional activity by the formation of a complex with both MyoD and MEF2 in the nucleus. Yeast-2 hybrid experiments illustrated PHB2’s direct interaction with MyoD and its indirect interaction with MEF2. These interactions were shown to be repressed by PHB2’s association with AKT2 (AKT serine/threonine kinase 2) [9][32]. Although this study illustrated that AKT2 interacts with PHB2, no modification of PHB2 by AKT2 was observed during myocytic differentiation [32]. Therefore, it was concluded that the disruption of the PHB2: MyoD: MEF2 interaction is not correlated to AKT2’s catalytic function [32].
A cotranscriptional activator of PPARγ, PGC-1α, has also been shown to interact with PHB2. This interaction inhibits the transcriptional activity of PPARγ [30]. PPARγ functions as a transcriptional regulator of many genes involved in activating adipogenesis. PHB2 levels are increased during adipogenesis; if PHB2 is knocked down, inhibition of adipogenesis is observed [9].
References
- McClung, J.K.; Danner, D.B.; Stewart, D.A.; Smith, J.R.; Schneider, E.L.; Lumpkin, C.K.; Dell’Orco, R.T.; Nuell, M.J. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem. Biophys. Res. Commun. 1989, 164, 1316–1322.
- Sato, T.; Saito, H.; Swensen, J.; Olifant, A.; Wood, C.; Danner, D.; Sakamoto, T.; Takita, K.; Kasumi, F.; Miki, Y.; et al. The human prohibitin gene located on chromosome 17q21 is mutated in sporadic breast cancer. Cancer Res. 1992, 52, 1643–1646.
- Nuell, M.J.; Stewart, D.A.; Walker, L.; Friedman, V.; Wood, C.M.; Owens, G.A.; Smith, J.R.; Schneider, E.L.; Dell’Orco, R.; Lumpkin, C.K.; et al. Prohibitin, an evolutionarily conserved intracellular protein that blocks DNA synthesis in normal fibroblasts and HeLa cells. Mol. Cell. Biol. 1991, 11, 1372–1381.
- Terashima, M.; Kim, K.M.; Adachi, T.; Nielsen, P.J.; Reth, M.; Kohler, G.; Lamers, M.C. The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. EMBO J. 1994, 13, 3782–3792.
- Montano, M.M.; Ekena, K.; Delage-Mourroux, R.; Chang, W.; Martini, P.; Katzenellenbogen, B.S. An estrogen receptor-selective coregulator that potentiates the effectiveness of antiestrogens and represses the activity of estrogens. Proc. Natl. Acad. Sci. USA 1999, 96, 6947–6952.
- Browman, D.T.; Hoegg, M.B.; Robbins, S.M. The SPFH domain-containing proteins: More than lipid raft markers. Trends Cell. Biol. 2007, 17, 394–402.
- Signorile, A.; Sgaramella, G.; Bellomo, F.; De Rasmo, D. Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells 2019, 8, 71.
- Tatsuta, T.; Model, K.; Langer, T. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 2005, 16, 248–259.
- Bavelloni, A.; Piazzi, M.; Raffini, M.; Faenza, I.; Blalock, W.L. Prohibitin 2: At a communications crossroads. IUBMB Life 2015, 67, 239–254.
- Funk, L.; Su, K.C.; Ly, J.; Feldman, D.; Singh, A.; Moodie, B.; Blainey, P.C.; Cheeseman, I.M. The phenotypic landscape of essential human genes. Cell 2022, 185, 4634–4653 e4622.
- Yang, J.; Li, B.; He, Q.Y. Significance of prohibitin domain family in tumorigenesis and its implication in cancer diagnosis and treatment. Cell Death Dis. 2018, 9, 580.
- Supale, S.; Thorel, F.; Merkwirth, C.; Gjinovci, A.; Herrera, P.L.; Scorrano, L.; Meda, P.; Langer, T.; Maechler, P. Loss of prohibitin induces mitochondrial damages altering beta-cell function and survival and is responsible for gradual diabetes development. Diabetes 2013, 62, 3488–3499.
- Wu, D.; Jian, C.; Peng, Q.; Hou, T.; Wu, K.; Shang, B.; Zhao, M.; Wang, Y.; Zheng, W.; Ma, Q.; et al. Prohibitin 2 deficiency impairs cardiac fatty acid oxidation and causes heart failure. Cell Death Dis. 2020, 11, 181.
- Merkwirth, C.; Martinelli, P.; Korwitz, A.; Morbin, M.; Bronneke, H.S.; Jordan, S.D.; Rugarli, E.I.; Langer, T. Loss of prohibitin membrane scaffolds impairs mitochondrial architecture and leads to tau hyperphosphorylation and neurodegeneration. PLoS Genet. 2012, 8, e1003021.
- Li, L.; Martin-Levilain, J.; Jimenez-Sanchez, C.; Karaca, M.; Foti, M.; Martinou, J.C.; Maechler, P. In vivo stabilization of OPA1 in hepatocytes potentiates mitochondrial respiration and gluconeogenesis in a prohibitin-dependent way. J. Biol. Chem. 2019, 294, 12581–12598.
- Chowdhury, I.; Thompson, W.E.; Thomas, K. Prohibitins role in cellular survival through Ras-Raf-MEK-ERK pathway. J. Cell Physiol. 2014, 229, 998–1004.
- Osman, C.; Merkwirth, C.; Langer, T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 2009, 122, 3823–3830.
- Zhou, Z.; Ai, H.; Li, K.; Yao, X.; Zhu, W.; Liu, L.; Yu, C.; Song, Z.; Bao, Y.; Huang, Y.; et al. Prohibitin 2 localizes in nucleolus to regulate ribosomal RNA transcription and facilitate cell proliferation in RD cells. Sci. Rep. 2018, 8, 1479.
- Ding, W.X.; Yin, X.M. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012, 393, 547–564.
- Ma, K.; Chen, G.; Li, W.; Kepp, O.; Zhu, Y.; Chen, Q. Mitophagy, Mitochondrial Homeostasis, and Cell Fate. Front. Cell Dev. Biol. 2020, 8, 467.
- Wei, Y.; Chiang, W.C.; Sumpter, R., Jr.; Mishra, P.; Levine, B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 2017, 168, 224–238.e210.
- Lahiri, V.; Klionsky, D.J. PHB2/prohibitin 2: An inner membrane mitophagy receptor. Cell Res. 2017, 27, 311–312.
- Yan, C.; Gong, L.; Chen, L.; Xu, M.; Abou-Hamdan, H.; Tang, M.; Desaubry, L.; Song, Z. PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy 2020, 16, 419–434.
- Peng, Y.T.; Chen, P.; Ouyang, R.Y.; Song, L. Multifaceted role of prohibitin in cell survival and apoptosis. Apoptosis 2015, 20, 1135–1149.
- Badrinath, N.; Yoo, S.Y. Mitochondria in cancer: In the aspects of tumorigenesis and targeted therapy. Carcinogenesis 2018, 39, 1419–1430.
- Merkwirth, C.; Dargazanli, S.; Tatsuta, T.; Geimer, S.; Lower, B.; Wunderlich, F.T.; von Kleist-Retzow, J.C.; Waisman, A.; Westermann, B.; Langer, T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008, 22, 476–488.
- Estaquier, J.; Vallette, F.; Vayssiere, J.L.; Mignotte, B. The mitochondrial pathways of apoptosis. Adv. Exp. Med. Biol. 2012, 942, 157–183.
- Bertolin, G.; Alves-Guerra, M.C.; Cheron, A.; Burel, A.; Prigent, C.; Le Borgne, R.; Tramier, M. Mitochondrial Aurora kinase A induces mitophagy by interacting with MAP1LC3 and Prohibitin 2. Life Sci. Alliance 2021, 4, e202000806.
- Wang, S.; Faller, D.V. Roles of prohibitin in growth control and tumor suppression in human cancers. Transl. Oncogenomics 2008, 3, 23–37.
- Thuaud, F.; Ribeiro, N.; Nebigil, C.G.; Desaubry, L. Prohibitin ligands in cell death and survival: Mode of action and therapeutic potential. Chem. Biol. 2013, 20, 316–331.
- Kasashima, K.; Ohta, E.; Kagawa, Y.; Endo, H. Mitochondrial functions and estrogen receptor-dependent nuclear translocation of pleiotropic human prohibitin 2. J. Biol. Chem. 2006, 281, 36401–36410.
- Sun, L.; Liu, L.; Yang, X.J.; Wu, Z. Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J. Cell Sci. 2004, 117, 3021–3029.
- Kurtev, V.; Margueron, R.; Kroboth, K.; Ogris, E.; Cavailles, V.; Seiser, C. Transcriptional regulation by the repressor of estrogen receptor activity via recruitment of histone deacetylases. J. Biol. Chem. 2004, 279, 24834–24843.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
735
Revisions:
2 times
(View History)
Update Date:
04 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




