
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mohammed Elmogy | -- | 2256 | 2022-12-06 19:35:15 | | | |
| 2 | Dean Liu | -14 word(s) | 2242 | 2022-12-07 02:05:46 | | |
Video Upload Options
Researchers presents a comprehensive survey on the diagnosis of colon cancer. This covers many aspects related to colon cancer, such as its symptoms and grades as well as the available imaging modalities (particularly, histopathology images used for analysis) in addition to common diagnosis systems. Furthermore, the most widely used datasets and performance evaluation metrics are discussed. Researchers provide a comprehensive review of the current studies on colon cancer, classified into deep-learning (DL) and machine-learning (ML) techniques, and researchers identify their main strengths and limitations. These techniques provide extensive support for identifying the early stages of cancer that lead to early treatment of the disease and produce a lower mortality rate compared with the rate produced after symptoms develop. In addition, these methods can help to prevent colorectal cancer from progressing through the removal of pre-malignant polyps, which can be achieved using screening tests to make the disease easier to diagnose. Finally, the existing challenges and future research directions that open the way for future work in this field are presented.
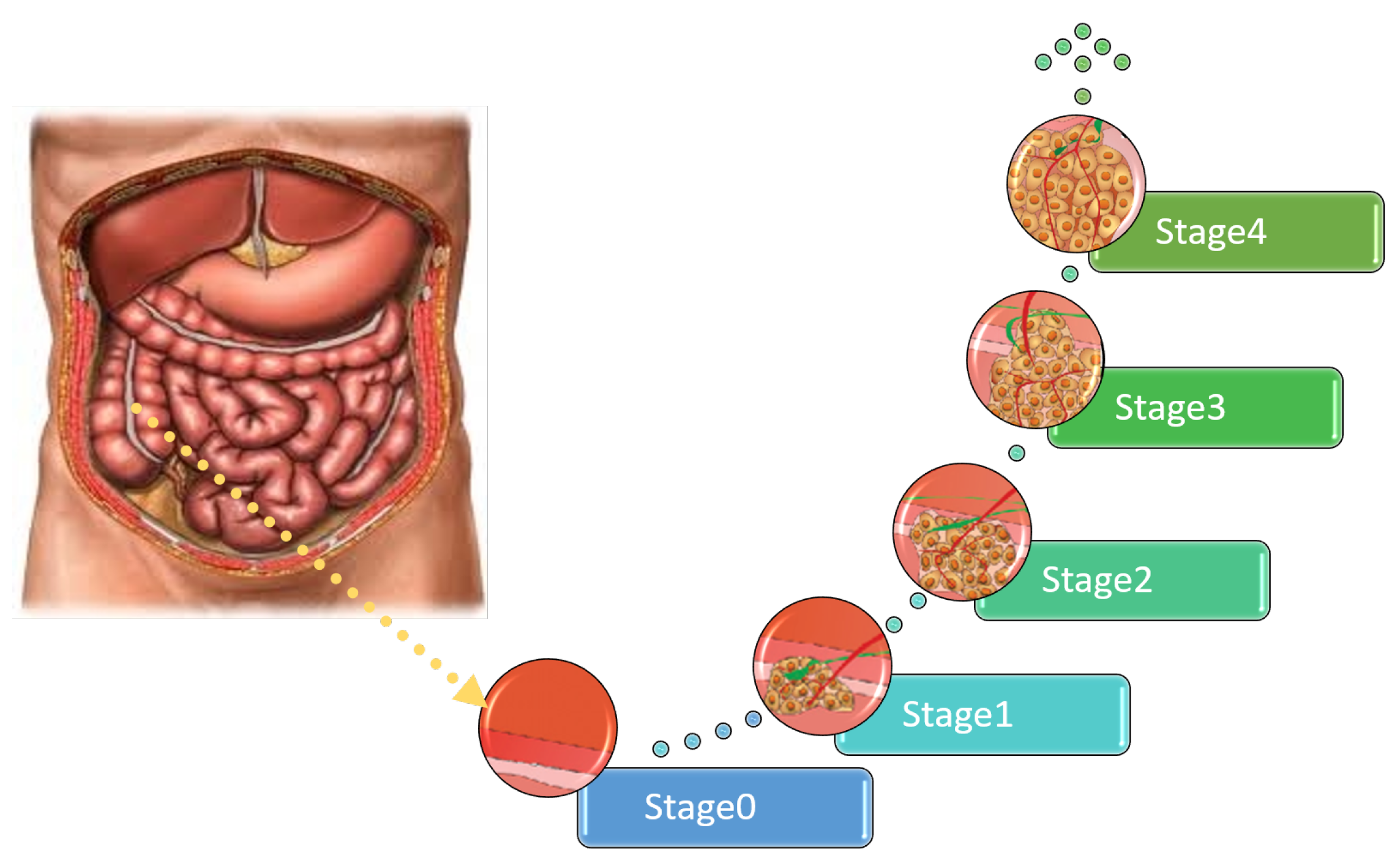
1. Introduction
| No. | Spread to Various Organs | Symptoms |
|---|---|---|
| 1 | Liver |
|
| 2 | Lung |
|
| 3 | Bone |
|
| 4 | Lymph nodes |
|
2. Colon Cancer Diagnosis
2.1. Imaging Modalities
2.2. Common Diagnosis Systems Based on HI Analysis

2.3. Datasets
-
CRC Grading DatasetThe CRC [26] Grading Dataset contains 38 H&E stained histological WSIs with a resolution 4548 × 7548.
-
PanNuke DatasetPanNuke [27] includes 200,000 nuclei divided into five main classes to challenge the approaches of classifying and segmenting nuclei in WSIs with a resolution of 224 × 224.
-
The Warwick-QU DatasetIn this dataset [28] are 16 slides of H&E stained histological WSIs of colon histology; this dataset is being created as category of the GlaS challenge with resolutions of 430 × 575 (14 images) and 520 × 775 (151 images).
-
CoNSeP DatasetCoNSeP [29] contains 41 H&E stained image slides with a resolution of 1000 × 1000 pixels at 40× magnification of objective: generally 24,319 annotated nuclei with labeled classes.
-
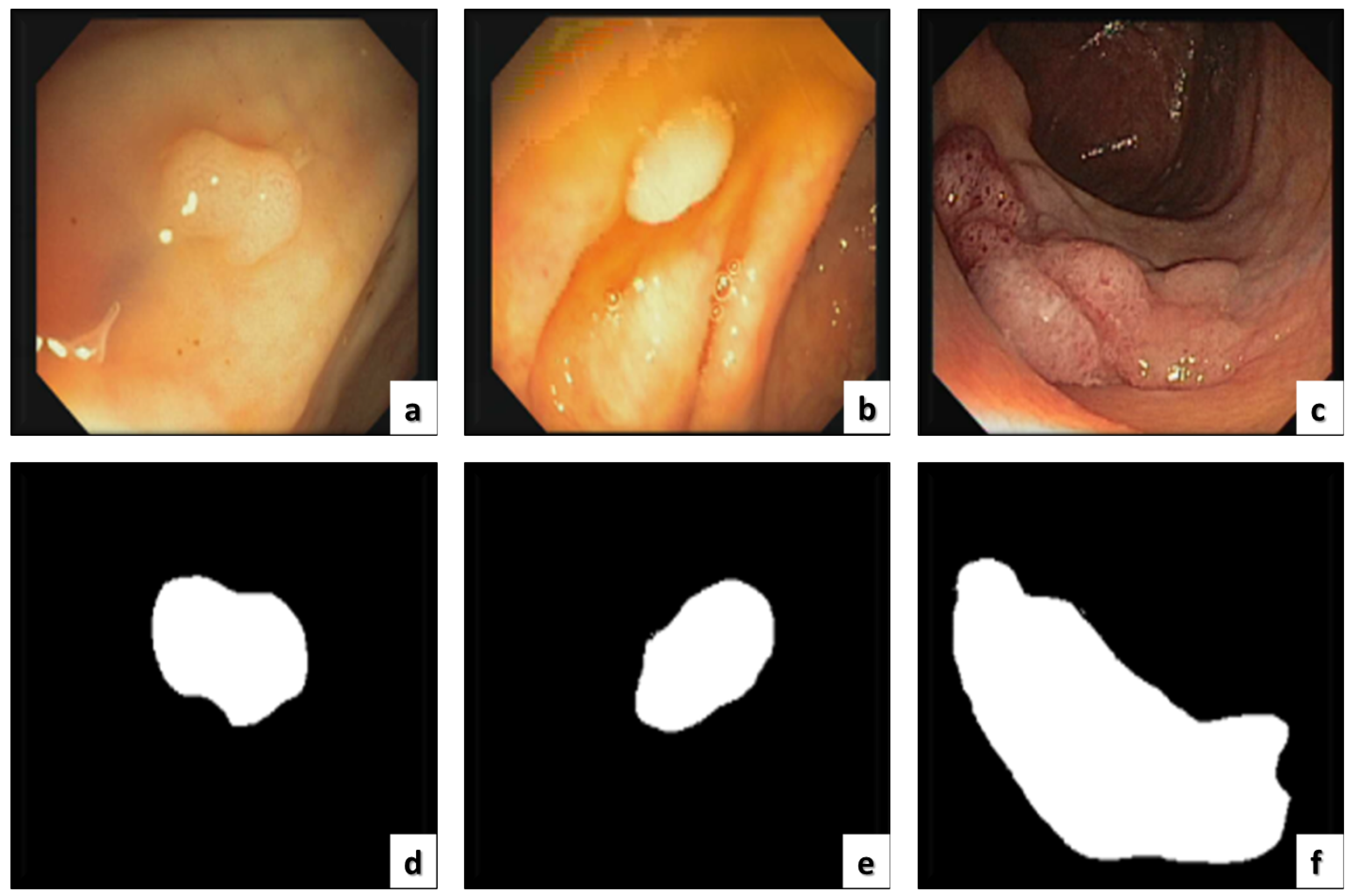
ETIS-LARIBThe ETIS-LARIB [30] database contains frames taken from colonoscopy videos, including several examples of polyps. It produces the baseline reality for each frame while displaying a mask due to the polyp region in the image. A sample of this dataset is shown in Figure 3.
-
CRCHistoPhenotypes–Labeled Cell Nuclei DatasetThis dataset [31] has 100 H&E CRC. For the process of detection, there are 29,756 nuclei; for classification, 22,444 nuclei (miscellaneous, fibroblast, and epithelial); and 7312 unlabeled with a resolution of 500 × 500.
-
Kent Integrated Dataset (KID)The KID [32] is responsible for the health and welfare system for the entire population of Medway and Kent. This dataset is rich and unique for researchers seeking health and care on a large scale. This also provides an overview of the patient journey, care, and needs.
-
CVC-ColonDB and CVC-ClinicDBSince 2012 [30], this dataset has been the top research leader as it includes many databases that are public and available, and CVC-ColonDB is included, which specializes in colon cancer imaging containing the original images and the ground truth as shown in Figure 4.
-
Colonoscopy DatasetThe dataset [33] contains 76 videos, containing both WL and NBI. The database contains 40 adenomas with SD resolution of 768 × 576, 21 hyperplastic lesions, and 15 serrated adenomas.
-
Extended CRC Grading Dataset (KID) In this dataset [34] are 300 images that are non-overlapping. These were labeled by expert pathologists as high grade (Grade 3) tumors, low grade (Grade 2) tumors, or normal tissue (Grade 1) with a resolution of 4548 × 7548.
-
ASU-Mayo ClinicCurrently, there are numerous research programs based on co-funded acceleration, seed research, and team science grants [35]. This means that more than 20–30 cohorts of senior nursing students in their clinical training by Mayo Clinic nursing faculty on the Mayo campus are expected to be completed. Due to this effort and cooperation, the seed grant program has added joint, cutting-edge research collaborations, a host of dual degree opportunities, and others. In 2016 and in the summer of 2010, the relationships of the Mayo Clinic became enterprise-wide, and the ASU Alliance for Health Care was formed.

3.4. Performance Evaluation Metrics
-
The Accuracy measures the proportion of true observations to the number of samples measured, which can be calculated as:
-
The Rate of Error shows the proportion of inaccurate observations to the number of measured samples, which can be calculated as:
-
The Precision measures the true classified positive estimates of the total classified estimates in a correct category, which can be calculated as:
-
The Recall is employed for measuring the ratio of correct estimates that are correctly predicted. This can be calculated as:
-
The Specificity is presented for measuring the positive observations rate of false samples and can be calculated as:
-
The Sensitivity measures the number of correct samples that are classified as true and can be calculated as:
-
The ROC curve presents the ratio of false positives to the ratio of TPs by showing the performances of the possible threshold values used and can be calculated as:
References
- Allison, J.E. Colorectal cancer screening guidelines: The importance of evidence and transparency. Gastroenterology 2010, 138, 1648–1652.
- An, F.P.; Liu, J.E. Medical Image Segmentation Algorithm Based on Optimized Convolutional Neural Network-Adaptive Dropout Depth Calculation. Complexity 2020, 2020, 1645479.
- Araghi, M.; Soerjomataram, I.; Jenkins, M.; Brierley, J.; Morris, E.; Bray, F.; Arnold, M. Global trends in colorectal cancer mortality: Projections to the year 2035. Int. J. Cancer 2019, 144, 2992–3000.
- Barish, M.A.; Soto, J.A.; Ferrucci, J.T. Consensus on current clinical practice of virtual colonoscopy. Am. J. Roentgenol. 2005, 184, 786–792.
- Thun, M.J.; Calle, E.E.; Namboodiri, M.M.; Flanders, W.D.; Coates, R.J.; Byers, T.; Boffetta, P.; Garfinkel, L.; Heath, C.W., Jr. Risk factors for fatal colon cancer in a large prospective study. JNCI J. Natl. Cancer Inst. 1992, 84, 1491–1500.
- Rathore, S.; Hussain, M.; Ali, A.; Khan, A. A recent survey on colon cancer detection techniques. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013, 10, 545–563.
- Baxter, N.N.; Goldwasser, M.A.; Paszat, L.F.; Saskin, R.; Urbach, D.R.; Rabeneck, L. Association of colonoscopy and death from colorectal cancer. Ann. Intern. Med. 2009, 150, 1–8.
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715.
- Kitayama, J.; Nagawa, H.; Tsuno, N.; Osada, T.; Hatano, K.; Sunami, E.; Saito, H.; Muto, T. Laminin mediates tethering and spreading of colon cancer cells in physiological shear flow. Br. J. Cancer 1999, 80, 1927–1934.
- Burdan, F.; Sudol-Szopinska, I.; Staroslawska, E.; Kolodziejczak, M.; Klepacz, R.; Mocarska, A.; Caban, M.; Zelazowska-Cieslinska, I.; Szumilo, J. Magnetic resonance imaging and endorectal ultrasound for diagnosis of rectal lesions. Eur. J. Med. Res. 2015, 20, 1–14.
- Bychkov, D.; Linder, N.; Turkki, R.; Nordling, S.; Kovanen, P.E.; Verrill, C.; Walliander, M.; Lundin, M.; Haglund, C.; Lundin, J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 2018, 8, 3395.
- Levin, B.; Lieberman, D.A.; McFarland, B.; Andrews, K.S.; Brooks, D.; Bond, J.; Dash, C.; Giardiello, F.M.; Glick, S.; Johnson, D.; et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: A joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology 2008, 134, 1570–1595.
- Chaddad, A.; Tanougast, C.; Dandache, A.; Al Houseini, A.; Bouridane, A. Improving of colon cancer cells detection based on Haralick’s features on segmented histopathological images. In Proceedings of the 2011 IEEE International Conference on Computer Applications and Industrial Electronics (ICCAIE), Penang, Malaysia, 21–22 May 2011; pp. 87–90.
- Hur, C.; Chung, D.C.; Schoen, R.E.; Gazelle, G.S. The management of small polyps found by virtual colonoscopy: Results of a decision analysis. Clin. Gastroenterol. Hepatol. 2007, 5, 237–244.
- Gunduz-Demir, C.; Kandemir, M.; Tosun, A.B.; Sokmensuer, C. Automatic segmentation of colon glands using object-graphs. Med. Image Anal. 2010, 14, 1–12.
- Wang, D.; Foran, D.J.; Ren, J.; Zhong, H.; Kim, I.Y.; Qi, X. Exploring automatic prostate histopathology image gleason grading via local structure modeling. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milano, Italy, 25–29 August 2015; pp. 2649–2652.
- Dal Molin, M.; Matthaei, H.; Wu, J.; Blackford, A.; Debeljak, M.; Rezaee, N.; Wolfgang, C.L.; Butturini, G.; Salvia, R.; Bassi, C.; et al. Clinicopathological correlates of activating GNAS mutations in intraductal papillary mucinous neoplasm (IPMN) of the pancreas. Ann. Surg. Oncol. 2013, 20, 3802–3808.
- Masud, M.; Sikder, N.; Nahid, A.A.; Bairagi, A.K.; AlZain, M.A. A machine learning approach to diagnosing lung and colon cancer using a deep learning-based classification framework. Sensors 2021, 21, 748.
- Elazab, N.; Soliman, H.; El-Sappagh, S.; Islam, S.; Elmogy, M. Objective Diagnosis for Histopathological Images Based on Machine Learning Techniques: Classical Approaches and New Trends. Mathematics 2020, 8, 1863.
- Bar-Shalom, R.; Valdivia, A.Y.; Blaufox, M.D. PET imaging in oncology. Semin. Nucl. Med. 2000, 30, 150–185.
- Moroz, M.A.; Kochetkov, T.; Cai, S.; Wu, J.; Shamis, M.; Nair, J.; De Stanchina, E.; Serganova, I.; Schwartz, G.K.; Banerjee, D.; et al. Imaging colon cancer response following treatment with AZD1152: A preclinical analysis of fluoro-2-deoxyglucose and fluorothymidine imaging. Clin. Cancer Res. 2011, 17, 1099–1110.
- Kalkan, H.; Nap, M.; Duin, R.P.; Loog, M. Automated classification of local patches in colon histopathology. In Proceedings of the Proceedings of the 21st International Conference on Pattern Recognition (ICPR2012); Tsukuba, Japan, 11–15 November 2012, pp. 61–64.
- Greenspan, H.; Van Ginneken, B.; Summers, R.M. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging 2016, 35, 1153–1159.
- Burt, R.W. Strategies for colon cancer screening with considerations of cost and access to care. J. Natl. Compr. Cancer Netw. 2010, 8, 2–5.
- Fakoor, R.; Ladhak, F.; Nazi, A.; Huber, M. Using deep learning to enhance cancer diagnosis and classification. In Proceedings of the International Conference on Machine Learning, New York, NY, USA, 15–20 July 2013; Volume 28.
- Awan, R.; Sirinukunwattana, K.; Epstein, D.; Jefferyes, S.; Qidwai, U.; Aftab, Z.; Mujeeb, I.; Snead, D.; Rajpoot, N. Glandular morphometrics for objective grading of colorectal adenocarcinoma histology images. Sci. Rep. 2017, 7, 1–12.
- Gamper, J.; Koohbanani, N.A.; Benes, K.; Graham, S.; Jahanifar, M.; Khurram, S.A.; Azam, A.; Hewitt, K.; Rajpoot, N. Pannuke dataset extension, insights and baselines. arXiv 2020, arXiv:2003.10778.
- Sirinukunwattana, K.; Snead, D.R.; Rajpoot, N.M. A stochastic polygons model for glandular structures in colon histology images. IEEE Trans. Med. Imaging 2015, 34, 2366–2378.
- Graham, S.; Vu, Q.D.; Raza, S.E.A.; Azam, A.; Tsang, Y.W.; Kwak, J.T.; Rajpoot, N. Hover-net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image Anal. 2019, 58, 101563.
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; de Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-seg: A segmented polyp dataset. In Proceedings of the International Conference on Multimedia Modeling, Daejeon, Republic of Korea, 5–8 January 2020; pp. 451–462.
- Sirinukunwattana, K.; Raza, S.E.A.; Tsang, Y.W.; Snead, D.R.; Cree, I.A.; Rajpoot, N.M. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imaging 2016, 35, 1196–1206.
- Lewer, D.; Bourne, T.; George, A.; Abi-Aad, G.; Taylor, C.; George, J. Data Resource: The Kent Integrated Dataset (KID). Int. J. Popul. Data Sci. 2018, 3, 427.
- Mesejo, P.; Pizarro, D.; Abergel, A.; Rouquette, O.; Beorchia, S.; Poincloux, L.; Bartoli, A. Computer-aided classification of gastrointestinal lesions in regular colonoscopy. IEEE Trans. Med. Imaging 2016, 35, 2051–2063.
- Shaban, M.; Awan, R.; Fraz, M.M.; Azam, A.; Tsang, Y.W.; Snead, D.; Rajpoot, N.M. Context-aware convolutional neural network for grading of colorectal cancer histology images. IEEE Trans. Med. Imaging 2020, 39, 2395–2405.
- Pogorelov, K.; Randel, K.R.; Griwodz, C.; Eskeland, S.L.; de Lange, T.; Johansen, D.; Spampinato, C.; Dang-Nguyen, D.T.; Lux, M.; Schmidt, P.T.; et al. Kvasir: A multi-class image dataset for computer aided gastrointestinal disease detection. In Proceedings of the eighth ACM on Multimedia Systems Conference, Taipei, Taiwan, 20–23 June 2017; pp. 164–169.





