
Video Upload Options
The emergence of human-induced Pluripotent Stem Cells (hiPSCs) has dramatically improved the understanding of human developmental processes under normal and diseased conditions. The hiPSCs have been differentiated into various tissue-specific cells in vitro, and the advancement in three-dimensional (3D) culture has provided a possibility to generate those cells in an in vivo-like environment. Tissues with 3D structures can be generated using different approaches such as self-assembled organoids and tissue-engineering methods, such as bioprinting. Researchers are interested in studying the self-assembled organoids differentiated from hiPSCs, as they have the potential to recapitulate the in vivo developmental process and be used to model human development and congenital defects. Organoids of tissues such as those of the intestine and brain were developed many years ago, but heart organoids were not reported until recently.
1. Introduction
2. Heart fields Formation
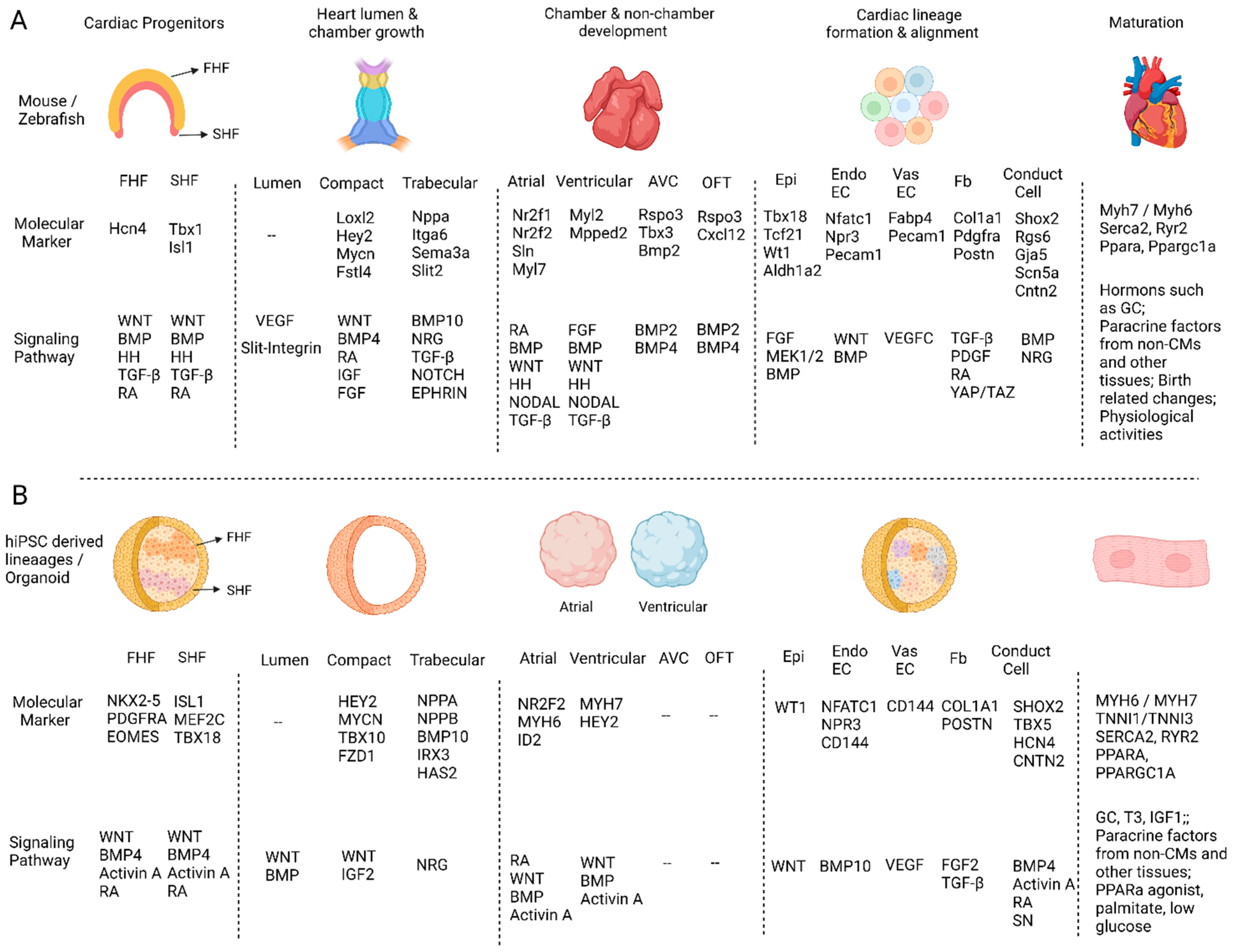
3. Heart Lumen Development
4. Compact and Trabecular Myocardium Growth
5. Heart Structure Development
References
- Mummery, C.L.; Zhang, J.; Ng, E.S.; Elliott, D.A.; Elefanty, A.G.; Kamp, T.J. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: A methods overview. Circ. Res. 2012, 111, 344–358.
- van den Brink, S.C.; Alemany, A.; van Batenburg, V.; Moris, N.; Blotenburg, M.; Vivié, J.; Baillie-Johnson, P.; Nichols, J.; Sonnen, K.F.; Martinez Arias, A. Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature 2020, 582, 405–409.
- Aguilera-Castrejon, A.; Oldak, B.; Shani, T.; Ghanem, N.; Itzkovich, C.; Slomovich, S.; Tarazi, S.; Bayerl, J.; Chugaeva, V.; Ayyash, M. Ex utero mouse embryogenesis from pre-gastrulation to late organogenesis. Nature 2021, 593, 119–124.
- Simunovic, M.; Brivanlou, A.H. Embryoids, organoids and gastruloids: New approaches to understanding embryogenesis. Development 2017, 144, 976–985.
- Sutherland, R.M.; McCredie, J.A.; Inch, W.R. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J. Natl. Cancer Inst. 1971, 46, 113–120.
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125.
- Sakalem, M.E.; De Sibio, M.T.; da Costa, F.A.d.S.; de Oliveira, M. Historical evolution of spheroids and organoids, and possibilities of use in life sciences and medicine. Biotechnol. J. 2021, 16, 2000463.
- Gilboa, S.M.; Devine, O.J.; Kucik, J.E.; Oster, M.E.; Riehle-Colarusso, T.; Nembhard, W.N.; Xu, P.; Correa, A.; Jenkins, K.; Marelli, A.J. Congenital heart defects in the United States: Estimating the magnitude of the affected population in 2010. Circulation 2016, 134, 101–109.
- Zou, Y.; Evans, S.; Chen, J.; Kuo, H.-C.; Harvey, R.P.; Chien, K.R. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development 1997, 124, 793–804.
- Buckingham, M.; Meilhac, S.; Zaffran, S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005, 6, 826–835.
- Liu, X.; Yagi, H.; Saeed, S.; Bais, A.S.; Gabriel, G.C.; Chen, Z.; Peterson, K.A.; Li, Y.; Schwartz, M.C.; Reynolds, W.T. The complex genetics of hypoplastic left heart syndrome. Nat. Genet. 2017, 49, 1152–1159.
- Andersen, P.; Tampakakis, E.; Jimenez, D.V.; Kannan, S.; Miyamoto, M.; Shin, H.K.; Saberi, A.; Murphy, S.; Sulistio, E.; Chelko, S.P. Precardiac organoids form two heart fields via Bmp/Wnt signaling. Nat. Commun. 2018, 9, 3140.
- Cai, C.-L.; Liang, X.; Shi, Y.; Chu, P.-H.; Pfaff, S.L.; Chen, J.; Evans, S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 2003, 5, 877–889.
- Azhar, M.; Schultz, J.E.J.; Grupp, I.; Dorn, I.I.G.W.; Meneton, P.; Molin, D.G.; Gittenberger-de Groot, A.C.; Doetschman, T. Transforming growth factor beta in cardiovascular development and function. Cytokine Growth Factor Rev. 2003, 14, 391–407.
- Kelly, R.G.; Buckingham, M.E.; Moorman, A.F. Heart fields and cardiac morphogenesis. Cold Spring Harb. Perspect. Med. 2014, 4, a015750.
- Rochais, F.; Mesbah, K.; Kelly, R.G. Signaling pathways controlling second heart field development. Circ. Res. 2009, 104, 933–942.
- Ryckebusch, L.; Wang, Z.; Bertrand, N.; Lin, S.-C.; Chi, X.; Schwartz, R.; Zaffran, S.; Niederreither, K. Retinoic acid deficiency alters second heart field formation. Proc. Natl. Acad. Sci. USA 2008, 105, 2913–2918.
- Knight, H.; Yelon, D. Utilizing zebrafish to understand second heart field development. Etiol. Morphog. Congenit. Heart Dis. 2016, 25, 193–199.
- Rossi, G.; Broguiere, N.; Miyamoto, M.; Boni, A.; Guiet, R.; Girgin, M.; Kelly, R.G.; Kwon, C.; Lutolf, M.P. Capturing cardiogenesis in gastruloids. Cell Stem Cell 2021, 28, 230–240.e6.
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Gabalski, M.A.; Volmert, B.D.; Ming, Y.; Ball, K.A.; Yang, W.; Zou, J.; Ni, G.; Pajares, N. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 2021, 12, 5142.
- Bertrand, N.; Roux, M.; Ryckebüsch, L.; Niederreither, K.; Dollé, P.; Moon, A.; Capecchi, M.; Zaffran, S. Hox genes define distinct progenitor sub-domains within the second heart field. Dev. Biol. 2011, 353, 266–274.
- Jammrath, J.; Reim, I.; Saumweber, H. Cbl-Associated Protein CAP contributes to correct formation and robust function of the Drosophila heart tube. PLoS ONE 2020, 15, e0233719.
- Knox, J.; Moyer, K.; Yacoub, N.; Soldaat, C.; Komosa, M.; Vassilieva, K.; Wilk, R.; Hu, J.; Paz, L.d.L.V.; Syed, Q. Syndecan contributes to heart cell specification and lumen formation during Drosophila cardiogenesis. Dev. Biol. 2011, 356, 279–290.
- Ivanovitch, K.; Temiño, S.; Torres, M. Live imaging of heart tube development in mouse reveals alternating phases of cardiac differentiation and morphogenesis. eLife 2017, 6, e30668.
- Strilić, B.; Kučera, T.; Eglinger, J.; Hughes, M.R.; McNagny, K.M.; Tsukita, S.; Dejana, E.; Ferrara, N.; Lammert, E. The molecular basis of vascular lumen formation in the developing mouse aorta. Dev. Cell 2009, 17, 505–515.
- Sydelko, B.S. Anatomy & Physiology Online. J. Med. Libr. Assoc. JMLA 2013, 101, 163.
- Hofbauer, P.; Jahnel, S.M.; Papai, N.; Giesshammer, M.; Deyett, A.; Schmidt, C.; Penc, M.; Tavernini, K.; Grdseloff, N.; Meledeth, C. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 2021, 184, 3299–3317.e22.
- Feng, W.; Schriever, H.; Jiang, S.; Bais, A.; Kostka, D.; Li, G. Computational profiling of hiPSC-derived heart organoids reveals chamber defects associated with Ebstein’s anomaly. bioRxiv 2020.
- Moorman, A.F.; Christoffels, V.M. Cardiac chamber formation: Development, genes, and evolution. Physiol. Rev. 2003, 83, 1223–1267.
- Schleich, J.-M.; Abdulla, T.; Summers, R.; Houyel, L. An overview of cardiac morphogenesis. Arch. Cardiovasc. Dis. 2013, 106, 612–623.
- Luxán, G.; D’Amato, G.; MacGrogan, D.; de la Pompa, J.L. Endocardial notch signaling in cardiac development and disease. Circ. Res. 2016, 118, e1–e18.
- Li, G.; Xu, A.; Sim, S.; Priest, J.R.; Tian, X.; Khan, T.; Quertermous, T.; Zhou, B.; Tsao, P.S.; Quake, S.R. Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev. Cell 2016, 39, 491–507.
- Li, G.; Tian, L.; Goodyer, W.; Kort, E.J.; Buikema, J.W.; Xu, A.; Wu, J.C.; Jovinge, S.; Wu, S.M. Single cell expression analysis reveals anatomical and cell cycle-dependent transcriptional shifts during heart development. Development 2019, 146, dev173476.
- Smith, T.K.; Bader, D.M. Signals from both sides: Control of cardiac development by the endocardium and epicardium. Semin. Cell Dev. Biol. 2007, 18, 84–89.
- Kang, J.-O.; Sucov, H.M. Convergent proliferative response and divergent morphogenic pathways induced by epicardial and endocardial signaling in fetal heart development. Mech. Dev. 2005, 122, 57–65.
- Díaz del Moral, S.; Benaouicha, M.; Muñoz-Chápuli, R.; Carmona, R. The Insulin-like Growth Factor Signalling Pathway in Cardiac Development and Regeneration. Int. J. Mol. Sci. 2022, 23, 234.
- Buikema, J.W.; Mady, A.S.; Mittal, N.V.; Atmanli, A.; Caron, L.; Doevendans, P.A.; Sluijter, J.P.; Domian, I.J. Wnt/β-catenin signaling directs the regional expansion of first and second heart field-derived ventricular cardiomyocytes. Development 2013, 140, 4165–4176.
- Bruneau, B.G. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb. Perspect. Biol. 2013, 5, a008292.
- Kodo, K.O.S.; Jahanbani, F.; Termglinchan, V.; Hirono, K.; InanlooRahatloo, K.; Ebert, A.D.; Shukla, P.; Abilez, O.J.; Churko, J.M.; Karakikes, I. iPSC-derived cardiomyocytes reveal abnormal TGF-β signalling in left ventricular non-compaction cardiomyopathy. Nat. Cell Biol. 2016, 18, 1031–1042.
- Chen, H.; Shi, S.; Acosta, L.; Li, W.; Lu, J.; Bao, S.; Chen, Z.; Yang, Z.; Schneider, M.D.; Chien, K.R. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 2004, 131, 2219–2231.
- Funakoshi, S.; Fernandes, I.; Mastikhina, O.; Wilkinson, D.; Tran, T.; Dhahri, W.; Mazine, A.; Yang, D.; Burnett, B.; Lee, J. Generation of mature compact ventricular cardiomyocytes from human pluripotent stem cells. Nat. Commun. 2021, 12, 3155.
- Mikryukov, A.A.; Mazine, A.; Wei, B.; Yang, D.; Miao, Y.; Gu, M.; Keller, G.M. BMP10 signaling promotes the development of endocardial cells from human pluripotent stem cell-derived cardiovascular progenitors. Cell Stem Cell 2021, 28, 96–111.e7.
- Lin, C.-J.; Lin, C.-Y.; Chen, C.-H.; Zhou, B.; Chang, C.-P. Partitioning the heart: Mechanisms of cardiac septation and valve development. Development 2012, 139, 3277–3299.
- Consortium, T.M. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 2018, 562, 367–372.
- Hochgreb, T.; Linhares, V.L.; Menezes, D.C.; Sampaio, A.C.; Yan, C.Y.; Cardoso, W.V.; Rosenthal, N.; Xavier-Neto, J. A caudorostral wave of RALDH2 conveys anteroposterior information to the cardiac field. Development 2003, 130, 5363–5374.
- Xavier-Neto, J.; Neville, C.M.; Shapiro, M.D.; Houghton, L.; Wang, G.F.; Nikovits, W.; Stockdale, F.E.; Rosenthal, N. A retinoic acid-inducible transgenic marker of sino-atrial development in the mouse heart. Development 1999, 126, 2677–2687.
- Yutzey, K.E.; Gannon, M.; Bader, D. Diversification of cardiomyogenic cell lineages in vitro. Dev. Biol. 1995, 170, 531–541.
- Martin, K.E.; Waxman, J.S. Atrial and sinoatrial node development in the zebrafish heart. J. Cardiovasc. Dev. Dis. 2021, 8, 15.
- O’Donnell, A.; Yutzey, K.E. Mechanisms of heart valve development and disease. Development 2020, 147, dev183020.
- Lee, J.H.; Protze, S.I.; Laksman, Z.; Backx, P.H.; Keller, G.M. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell 2017, 21, 179–194.e4.
- Cyganek, L.; Tiburcy, M.; Sekeres, K.; Gerstenberg, K.; Bohnenberger, H.; Lenz, C.; Henze, S.; Stauske, M.; Salinas, G.; Zimmermann, W.-H. Deep phenotyping of human induced pluripotent stem cell–derived atrial and ventricular cardiomyocytes. JCI Insight 2018, 3, e99941.
- Friedman, C.E.; Nguyen, Q.; Lukowski, S.W.; Helfer, A.; Chiu, H.S.; Miklas, J.; Levy, S.; Suo, S.; Han, J.-D.J.; Osteil, P. Single-cell transcriptomic analysis of cardiac differentiation from human PSCs reveals HOPX-dependent cardiomyocyte maturation. Cell Stem Cell 2018, 23, 586–598.e8.
- Grancharova, T.; Gerbin, K.A.; Rosenberg, A.B.; Roco, C.M.; Arakaki, J.E.; DeLizo, C.M.; Dinh, S.Q.; Donovan-Maiye, R.M.; Hirano, M.; Nelson, A.M. A comprehensive analysis of gene expression changes in a high replicate and open-source dataset of differentiating hiPSC-derived cardiomyocytes. Sci. Rep. 2021, 11, 15845.
- Kathiriya, I.S.; Rao, K.S.; Iacono, G.; Devine, W.P.; Blair, A.P.; Hota, S.K.; Lai, M.H.; Garay, B.I.; Thomas, R.; Gong, H.Z. Modeling human TBX5 haploinsufficiency predicts regulatory networks for congenital heart disease. Dev. Cell 2021, 56, 292–309.e9.
- Paige, S.L.; Galdos, F.X.; Lee, S.; Chin, E.T.; Ranjbarvaziri, S.; Feyen, D.A.; Darsha, A.K.; Xu, S.; Ryan, J.A.; Beck, A.L. Patient-specific induced pluripotent stem cells implicate intrinsic impaired contractility in hypoplastic left heart syndrome. Circulation 2020, 142, 1605–1608.




