Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Shun Yaginuma | -- | 1640 | 2022-04-25 15:19:04 | | | |
| 2 | Camila Xu | + 322 word(s) | 1962 | 2022-04-26 03:54:58 | | | | |
| 3 | Camila Xu | Meta information modification | 1962 | 2022-04-26 04:05:41 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Yaginuma, S.; Aoki, J.; , . Phospholipase A1. Encyclopedia. Available online: https://encyclopedia.pub/entry/22246 (accessed on 02 March 2026).
Yaginuma S, Aoki J, . Phospholipase A1. Encyclopedia. Available at: https://encyclopedia.pub/entry/22246. Accessed March 02, 2026.
Yaginuma, Shun, Junken Aoki, . "Phospholipase A1" Encyclopedia, https://encyclopedia.pub/entry/22246 (accessed March 02, 2026).
Yaginuma, S., Aoki, J., & , . (2022, April 25). Phospholipase A1. In Encyclopedia. https://encyclopedia.pub/entry/22246
Yaginuma, Shun, et al. "Phospholipase A1." Encyclopedia. Web. 25 April, 2022.
Copy Citation
Phospholipase A1 (PLA1) is an enzyme that cleaves an ester bond at the sn-1 position of glycerophospholipids, producing a free fatty acid and a lysophospholipid.
phospholipase A1
phospholipid metabolism
lysophospholipid
1. Introduction
Phospholipase A1 (PLA1) is an enzyme that hydrolyzes an ester bond at the sn-1 position of glycerophospholipids (GPLs), usually producing a saturated or mono-unsaturated fatty acid and a 1-lyso-2-acyl-phospholipid (2-acyl-lysophospholipid, 2-acyl-LPL) (Figure 1). PLA1 has not attracted as much attention as other mammalian acyl hydrolases, such as phospholipase A2 (PLA2), which hydrolyzes fatty acids, mainly unsaturated fatty acids, and acts as a first step in producing enzymes for bioactive lipids such as eicosanoids and platelet-activating factor (PAF) [1][2]. Some PLA1s and PLA2 target neutral lipids such as triacylglycerol (TAG) and diacylglycerol (DAG) in addition to GPLs (Figure 1).
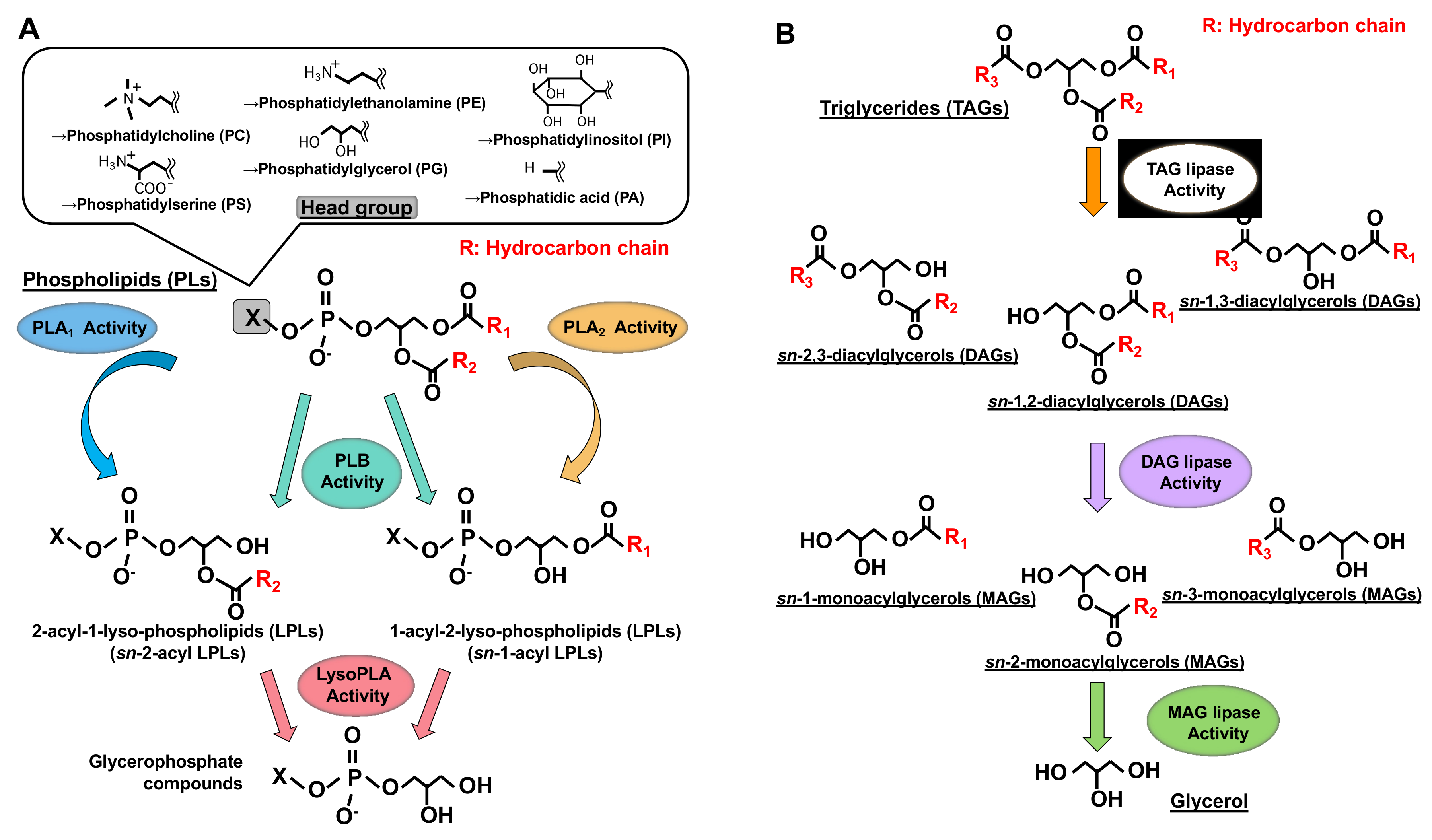 Figure 1. Structures of glycerolipids and their metabolic enzymes. (A) Glycerophospholipids (GPL) and phospholipases. GPLs are composed of a polar head group (six major classes), a glycerol backbone and fatty acid moieties (esterified at the sn-1 and sn-2 positions). Phospholipase A1 (PLA1) hydrolyzes a fatty acid at the sn-1 position, generating sn-2-acyl-1-lyso-phospholipids (sn-2-acyl LPLs), while phospholipase A2 (PLA2) hydrolyzes a fatty acid at the sn-2 position generating sn-1-acyl-2-lyso-phospholipids (sn-1-acyl LPLs). Phospholipase B (PLB) hydrolyzes a fatty acid at both sn-1 and sn-2 positions. LysoPLA hydrolyzes a fatty acid of sn-2-acyl LPLs and sn-1-acyl LPLs, generating glycerophosphate compounds. (B) Triacylglycerol (TAG) has three fatty acids at the sn-1, sn-2 and sn-3 positions of glycerol backbone, diacylglycerol (DAG) has two fatty acids and monoacylglycerol (MAG) has one fatty acid. TAG lipase hydrolyzes a fatty acid of TAG, generating sn-1, 2, sn-2, 3 or sn-1, 3-diacylglycerols (DAGs). DAG lipase hydrolyzes a fatty acid of DAG and MAG lipase hydrolyzes a fatty acid of MAG.
Figure 1. Structures of glycerolipids and their metabolic enzymes. (A) Glycerophospholipids (GPL) and phospholipases. GPLs are composed of a polar head group (six major classes), a glycerol backbone and fatty acid moieties (esterified at the sn-1 and sn-2 positions). Phospholipase A1 (PLA1) hydrolyzes a fatty acid at the sn-1 position, generating sn-2-acyl-1-lyso-phospholipids (sn-2-acyl LPLs), while phospholipase A2 (PLA2) hydrolyzes a fatty acid at the sn-2 position generating sn-1-acyl-2-lyso-phospholipids (sn-1-acyl LPLs). Phospholipase B (PLB) hydrolyzes a fatty acid at both sn-1 and sn-2 positions. LysoPLA hydrolyzes a fatty acid of sn-2-acyl LPLs and sn-1-acyl LPLs, generating glycerophosphate compounds. (B) Triacylglycerol (TAG) has three fatty acids at the sn-1, sn-2 and sn-3 positions of glycerol backbone, diacylglycerol (DAG) has two fatty acids and monoacylglycerol (MAG) has one fatty acid. TAG lipase hydrolyzes a fatty acid of TAG, generating sn-1, 2, sn-2, 3 or sn-1, 3-diacylglycerols (DAGs). DAG lipase hydrolyzes a fatty acid of DAG and MAG lipase hydrolyzes a fatty acid of MAG.Much is known about the functions of PLA2, whereas those of PLA1 remain limited. However, because fatty acids at both the sn-1 and sn-2 positions of GPLs have a high turnover rate [3], PLA1 as well as PLA2, appears to be involved in the rapid turnover and remodeling of cellular GPLs (Figure 2). In addition, some PLA1s also have a specific role in the production of 2-acyl-1-lysophospholipids, which serve as lysophospholipid mediators. For example, one type of PLA1, membrane-associated phosphatidic acid-selective PLA1 (mPA-PLA1α in Table 1, Figure 3A), produces 2-acyl-1-lysophosphatidic acid (2-acyl-lysoPA (LPA)) with an unsaturated fatty acid residue [4]. The 2-acyl-LPA acts as a potent ligand for LPAR3/EDG7 and LPAR6/P2Y5, with LPA receptors preferring 2-acyl-LPA over 1-acyl-LPA [5][6]. Phosphatidylserine-specific PLA1 (PS-PLA1 in Table 1, Figure 3A) also acts as a producing enzyme of another lysophospholipid mediator, 2-acyl-lysophosphatidylserine (2-acyl-lysoPS (LysoPS)), which further supports the idea that PLA1s function as producing enzymes for lysophospholipid mediators.
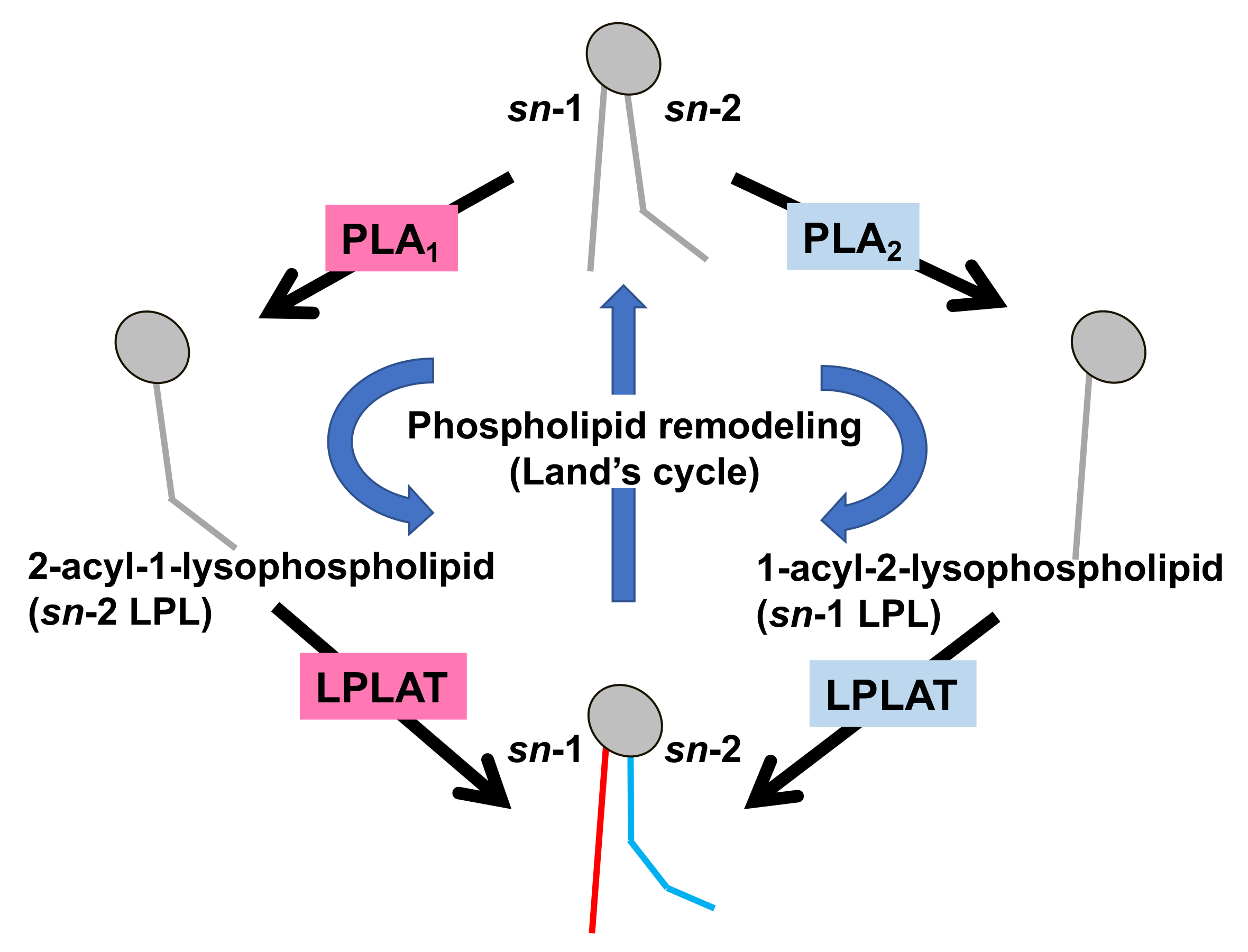 Figure 2. Fatty acid remodeling reactions of GPLs. Glycerophospholipids (GPL) in the cells are constantly subjected to two kinds of fatty acid hydrolyzing reactions mediated by phospholipase A1 (PLA1) and phospholipase A2 (PLA2), resulting in the production of 2-acyl-1-lysophospholipid (sn-2 LPL) and 1-acyl-2-lysophospholipid (sn-1 LPL). The LPLs thus produced are further subjected to acylation reactions to re-form the GPLs. Several kinds of lysophospholipid acyltransferases (LPLAT) are responsible for the introduction of fatty acids to lysophospholipids. By these sequential GPL remodeling reactions, the fatty acids of GPLs are constantly replaced.
Figure 2. Fatty acid remodeling reactions of GPLs. Glycerophospholipids (GPL) in the cells are constantly subjected to two kinds of fatty acid hydrolyzing reactions mediated by phospholipase A1 (PLA1) and phospholipase A2 (PLA2), resulting in the production of 2-acyl-1-lysophospholipid (sn-2 LPL) and 1-acyl-2-lysophospholipid (sn-1 LPL). The LPLs thus produced are further subjected to acylation reactions to re-form the GPLs. Several kinds of lysophospholipid acyltransferases (LPLAT) are responsible for the introduction of fatty acids to lysophospholipids. By these sequential GPL remodeling reactions, the fatty acids of GPLs are constantly replaced.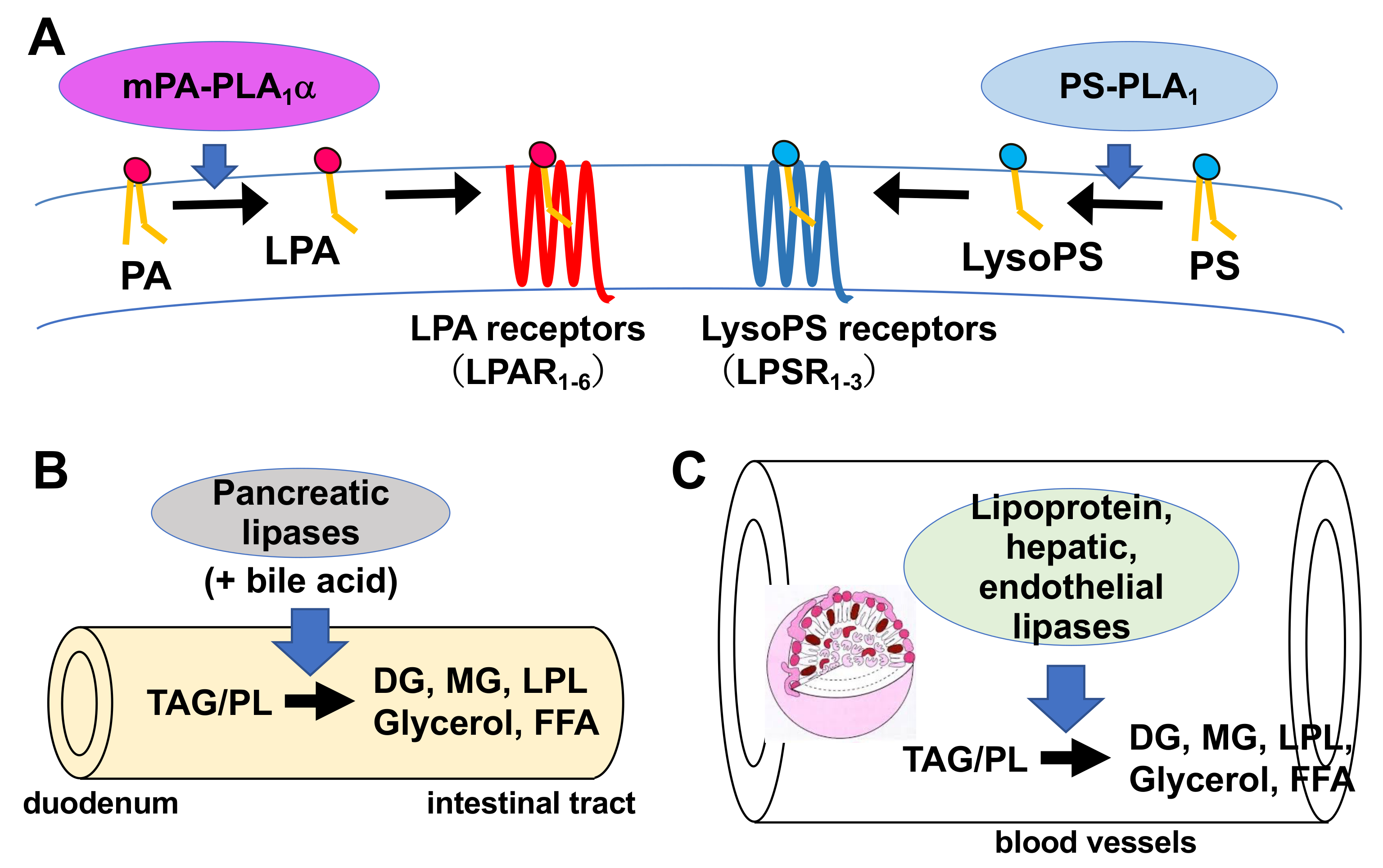 Figure 3. Physiological roles of extracellular PLA1s. (A) PS-PLA1 and mPA-PLA1α serve as producing enzymes for lysophospholipid mediators. PS-PLA1 has a strict substrate specificity in that it only acts on serine containing GPLs such as phosphatidylserine (PS) and lysophosphatidylserine (LysoPS). LysoPS then acts on GPCR-type LysoPS receptors. Three such LysoPS receptors have been identified. These include LPSR1/GPR34, LPSR2/P2Y10, and LPSR3/GPR174. mPA-PLA1α acts on PA in a specific manner and produces sn-2 LPA, which then acts on GPCR-type LPA receptors, LPAR1-LPAR6 evoking various biological responses. (B) Pancreatic lipase (PL) is secreted from the pancreas into the lumen of the intestine, where it, with the aid of bile acids, hydrolyzes the fatty acids of triacylglycerol (TAG) and GPLs in the digestive juice yielding diacylglycerol (DAG), monoacylglycerol (MAG) and fatty acids. The liberated fatty acids are absorbed by intestinal cells as nutrients. (C) Lipoprotein lipase (LPL), hepatic lipase (HL), and endothelial lipase (EL), which are mainly present in the blood, are associated with endothelial cell surfaces in adipose tissues (LPL), heart (LPL), liver (HL) and various tissues. These lipases have both TAG lipase and PLA1 activities. They hydrolyze fatty acids of TAG and GPLs present in the circulating lipoproteins such as low-density lipoproteins (LDL) and high-density lipoproteins (HDL), yielding diacylglycerol (DAG), monoacylglycerol (MAG), lysophospholipids (LPL) and fatty acids. The free fatty acids are absorbed by corresponding cells for energy source and storage of neutral lipids.
Figure 3. Physiological roles of extracellular PLA1s. (A) PS-PLA1 and mPA-PLA1α serve as producing enzymes for lysophospholipid mediators. PS-PLA1 has a strict substrate specificity in that it only acts on serine containing GPLs such as phosphatidylserine (PS) and lysophosphatidylserine (LysoPS). LysoPS then acts on GPCR-type LysoPS receptors. Three such LysoPS receptors have been identified. These include LPSR1/GPR34, LPSR2/P2Y10, and LPSR3/GPR174. mPA-PLA1α acts on PA in a specific manner and produces sn-2 LPA, which then acts on GPCR-type LPA receptors, LPAR1-LPAR6 evoking various biological responses. (B) Pancreatic lipase (PL) is secreted from the pancreas into the lumen of the intestine, where it, with the aid of bile acids, hydrolyzes the fatty acids of triacylglycerol (TAG) and GPLs in the digestive juice yielding diacylglycerol (DAG), monoacylglycerol (MAG) and fatty acids. The liberated fatty acids are absorbed by intestinal cells as nutrients. (C) Lipoprotein lipase (LPL), hepatic lipase (HL), and endothelial lipase (EL), which are mainly present in the blood, are associated with endothelial cell surfaces in adipose tissues (LPL), heart (LPL), liver (HL) and various tissues. These lipases have both TAG lipase and PLA1 activities. They hydrolyze fatty acids of TAG and GPLs present in the circulating lipoproteins such as low-density lipoproteins (LDL) and high-density lipoproteins (HDL), yielding diacylglycerol (DAG), monoacylglycerol (MAG), lysophospholipids (LPL) and fatty acids. The free fatty acids are absorbed by corresponding cells for energy source and storage of neutral lipids.Table 1. Mammalian PLA1s.
| Primary Name | Other Name | Human Gene | Substrate | Reaction Mediated | PDB ID (H: Human, R: Rat) |
Ref. | |
|---|---|---|---|---|---|---|---|
| extracellular PLA1s | PS-PLA1 | PLA1A | PLA1A | PS | Producing enzyme for bioactive lysophospholipid, LysoPS | - | [7][8] |
| PA-PLA1α | LIPH, mPA-PLA1α | LIPH | PA | Producing enzyme for bioactive lysophospholipid, LPA | - | [4][9] | |
| lipoprotein lipase | LPL, LIPD | LIPD | TAG, PL | TAG lipase and PLA1 activity | H: 6E7K, 6OAU, 6OAZ, 6OB0, 6WN4 | [10] | |
| hepatic lipase | HL, LIPC | LIPC | TAG, PL | TAG lipase and PLA1 activity | - | [10] | |
| endothelial cell-derived lipase | EDL, EL, LIPG | LIPG | PL | Predominant PLA1 activity | - | ||
| pacreatic lipase | PL, PNLIP | PNLIP | TAG, PL | TAG lipase and PLA1 activity | H: 1GPL, 1LPA, 1LPB, 1N8S | [11][12][13] | |
| pancreatic lipase-related protein 1 | PLRP1 | PLRP1 | TAG, PL | TAG lipase and PLA1 activity | H: 2PPL | ||
| pancreatic lipase-related protein 2 | PLRP2 | PLRP2 | TAG, PL | TAG lipase and PLA1 activity | H: 2OXE, 2PVS; R: 1BU8 | [11][13] | |
| intracellular PLA1s | PA-PLA1 | DDHD1, iPLA1α | DDHD1 | PL | PLA1 activity | - | [14][15] |
| KIAA0725p | DDHD2, iPLA1γ | DDHD2 | PL | PE, DAG, CL | - | [16][17][18] | |
| p125 | iPLA1β | P125 | n.d. | Enzymatic activity has not been detected | - | [19] | |
| PNPLA6 | iPLA2δ, NTE | PNPLA6 | PC, LPC | PLB, LysoPLA activity cleaving FAs at both sn-1 and sn-2 positions | - | [20][21][22] | |
| PNPLA7 | iPLA2θ, NRE | PNPLA7 | PC, LPC | PLB, LysoPLA activity cleaving FAs at both sn-1 and sn-2 positions | - | [23] | |
| PNPLA8 | iPLA2γ, Group VIB PLA2 | PNPLA8 | PC | PLB activity cleaving FAs at both sn-1 and sn-2 positions | - | [24][25] | |
| cPLA2α | PLA2G4A, Group IVA PLA2 | PLA2G4A | PL | PLB activity cleaving FAs at both sn-1 and sn-2 positions | H: 1BCI, 1CJY, 1RLW | [26] | |
| cPLA2β | PLA2G4B, Group IVB PLA2 | PLA2G4B | PL | PLB activity cleaving FAs at both sn-1 and sn-2 positions | - | [26][27] | |
| cPLA2ζ | PLA2G4F, Group IVF PLA2 | PLA2G4F | PL | PLB activity cleaving FAs at both sn-1 and sn-2 positions | - | ||
| PLA2G16 | Group XVI PLA2, PLAAT3, HRASLS3, H-Rev107 | PLA2G16 | PL | PLB activity cleaving FAs at both sn-1 and sn-2 positions | H: 2KYT, 4DOT, 4FA0, 4Q95, 7C3Z, 7C41 | [28] |
Although PLA1 activity has been detected in many mammalian tissues and cells [29][30][31][32][33][34][35][36], only a few PLA1s have been purified and cloned. Some triacylglycerol (TAG)-hydrolyzing lipases (Figure 1) such as hepatic lipase, endothelial cell lipase, and lipoprotein lipase also show PLA1 activity [11].
2. History of PLA1 Research
The following is a brief history of mammalian PLA1 research. PLA1 as well as PLA2 activities have been detected in various tissues and plasma. In the 1990’s, two PLA1 molecules were biochemically purified and identified. These two PLA1s are phosphatidic acid-preferential PLA1 (PA-PLA1) and phosphatidylserine-specific PLA1 (PS-PLA1) (Table 1). In 1994, Higgs and Glomset purified a novel PLA1 from a cytosolic fraction of bovine testes that preferentially hydrolyzed PA [14] (Table 1). Shortly after, the same group cloned a cDNA of the PLA1, and PA-PLA1 was found to be an intracellular protein composed of 875-amino acids with a calculated molecular mass of approximately 100 kDa. Simultaneously, Exton’s group purified a very similar PLA1 from the bovine brain [37]. It was not clear whether the two intracellular PLA1s were identical, because the two groups used different PLA1 assay conditions and substrates. Later, Kudo’s group purified similar PLA1s from the brain and testes of mice, rats, and cows [38] and showed that they were identical to the PA-PLA1 mentioned above. They demonstrated that PA-PLA1 was predominantly hydrolyzed phosphatidic acid (PA) in the presence of Triton X-100 and phosphatidylethanolamine (PE) in its absence. The result showed clearly that the substrate specificity of PA-PLA1 in vitro is affected by the assay conditions, which makes it difficult to identify the natural substrates of PA-PLA1. This also implies that the name, PA-PLA1, is not suitable for the enzyme.
Horigome et al., detected two PLA activities in the supernatant of activated rat platelets [39][40]. One was identified as secretory PLA2, now known as Group IIA secretory phospholipase A2 (sPLA2IIA). Sato et al., succeeded in purifying and cloning another PLA that showed a high preference for serine-containing phospholipids [7]. The PLA is now known as PS-specific PLA1 (PS-PLA1) (Table 1). PS-PLA1 specifically hydrolyzed PS in vitro. In addition, it acted on the intact cell membrane, hydrolyzed PS and produced 2-acyl-LysoPS. Thus, PS-PLA1 is believed to be a LysoPS-producing enzyme.
After discovering the two PLA1 molecules, several researchers found similar PLA1s (homologs) in the nucleotide databases, expressed them as recombinant proteins and characterized them biochemically. These analyses identified the homologs as novel PLA1s. These include extracellular enzyme membrane-associated PA-selective PLA1α (mPA-PLA1α, a PS-PLA1 homologue) [4] (Table 1) and intracellular enzyme iPLA1γ/DDHD2/KIAA0725 (a PA-PLA1 homologue) [16] and iPLA1β/SEC23IP/p125 (PLA1 activities have not been detected) [19] (Table 1). In addition, similar biochemical characterization was performed for PLA2 homologs. Interestingly, biochemical characterizations of the PLA2 homologs showed that some of them exhibited PLA1 activity in addition to their PLA2 activity (PLB activity (Figure 1)) (Table 1). It should also be noted here that certain lipases that hydrolyze TAG have PLA1 activity, as mentioned above (Table 1). Lipases hydrolyze fatty acids at the sn-1 and sn-3 positions of triacylglycerol (TAG) and diacylglycerol (DAG). They also hydrolyze fatty acids at the sn-1 or sn-3 positions of monoacylglycerol (MAG). This research will summarize current knowledge on PLA1 molecules reported thus far, discussing their discoveries, structures, tissue and cellular distributions, and possible biological functions.
PLA1s are roughly divided into two groups: (1) extracellular PLA1/lipase family and (2) intracellular PLA1 family (Table 1), depending on their cellular localization and primary amino acid sequences. The intracellular PLA1 family was further subdivided into several groups, consisting of an iPLA1 family, PNPLA family, cPLA2 family and PLAAT family.
3. Structural Evaluation of PLA1 Molecules
Recently, a computational machine learning method named AlphaFold was developed enabling researchers to predict protein structures with high accuracy, even when no similar experimentally solved structure is available [41]. In this research, researchers utilized AlphaFold to generate structural predictions of PLA1 molecules. Figure 4 summarizes the AlphaFold-generated structures of some PLA1 molecules focused on in this research. Note that some structures have been determined, e.g., by X-ray crystallography, while others were structure-predicted. Since structures of any PNPLA and iPLA1 family members have not been determined, AlphaFold was unable to predict their structures. Thus, predicted structures were shown only for extracellular PLA1/lipase and for cPLA2 family members (Figure 4).
 Figure 4. Structures of PLA1 molecules. 3D structures of three PLA1 family members (extracellular PLA1/lipase, cPLA2 and PLAAT family members) were shown. For extracellular PLA1/lipase family members, β5 and the β9 loops and the lid domain are shown in yellow, cyan and green, respectively. The three residues forming a catalytic triad (Ser, Asp and His) are described as sticks (red). For cPLA2 family members (cPLA2α, cPLA2β and cPLA2ζ), the conserved lipase motifs (GXSGX and DXG) are shown in red, and the two residues forming a catalytic dyad (Ser and Asp) are shown as sticks. For PLAAT3/PLA2G16, the three residues forming a catalytic triad (two histidines and cysteine) are shown as sticks (red). The structures without asterisk were acquired from RCSB Protein Data Bank. (Reference PDB IDs; PL (1LPB), LPL (6OB0), PLRP1 (2PPL), cPLA2α (1CJY), PLA2G16 (4DOT)). The predicted structures of lipases with asterisk were acquired from AlphaFold Protein Structure Database. All the structures were visualized using PyMOL software.
Figure 4. Structures of PLA1 molecules. 3D structures of three PLA1 family members (extracellular PLA1/lipase, cPLA2 and PLAAT family members) were shown. For extracellular PLA1/lipase family members, β5 and the β9 loops and the lid domain are shown in yellow, cyan and green, respectively. The three residues forming a catalytic triad (Ser, Asp and His) are described as sticks (red). For cPLA2 family members (cPLA2α, cPLA2β and cPLA2ζ), the conserved lipase motifs (GXSGX and DXG) are shown in red, and the two residues forming a catalytic dyad (Ser and Asp) are shown as sticks. For PLAAT3/PLA2G16, the three residues forming a catalytic triad (two histidines and cysteine) are shown as sticks (red). The structures without asterisk were acquired from RCSB Protein Data Bank. (Reference PDB IDs; PL (1LPB), LPL (6OB0), PLRP1 (2PPL), cPLA2α (1CJY), PLA2G16 (4DOT)). The predicted structures of lipases with asterisk were acquired from AlphaFold Protein Structure Database. All the structures were visualized using PyMOL software.References
- Dennis, E.A. The growing phospholipase A2 superfamily of signal transduction enzymes. Trends Biochem. Sci. 1997, 22, 1–2.
- Murakami, M.; Nakatani, Y.; Atsumi, G.; Inoue, K.; Kudo, I. Regulatory functions of phospholipase A2. Crit. Rev. Immunol. 1997, 17, 225–283.
- Lykidis, A.; Jackowski, S. Regulation of mammalian cell membrane biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 2001, 65, 361–393.
- Sonoda, H.; Aoki, J.; Hiramatsu, T.; Ishida, M.; Bandoh, K.; Nagai, Y.; Taguchi, R.; Inoue, K.; Arai, H. A novel phosphatidic acid-selective phospholipase A1 that produces lysophosphatidic acid. J. Biol. Chem. 2002, 277, 34254–34263.
- Bandoh, K.; Aoki, J.; Tsujimoto, M.; Arai, H.; Inoue, K. Lysophosphatidic acid (LPA) receptors of the EDG family are differentially activated by LPA species. Structure-activity relationship of cloned LPA receptors. FEBS Lett. 2000, 478, 159–165.
- Bandoh, K.; Aoki, J.; Hosono, H.; Kobayashi, S.; Kobayashi, T.; Murakami, M.K.; Tsujimoto, M.; Arai, H.; Inoue, K. Molecular cloning and characterization of a novel human G-protein-coupled receptor, EDG7, for lysophosphatidic acid. J. Biol. Chem. 1999, 274, 27776–27785.
- Sato, T.; Aoki, J.; Nagai, Y.; Dohmae, N.; Takio, K.; Doi, T.; Arai, H.; Inoue, K. Serine phospholipid-specific phospholipase A that is secreted from activated platelets. A new member of the lipase family. J. Biol. Chem. 1997, 272, 2192–2198.
- Nagai, Y.; Aoki, J.; Sato, T.; Amano, K.; Matsuda, Y.; Arai, H.; Inoue, K. An alternative splicing form of phosphatidylserine-specific phospholipase A1 that exhibits lysophosphatidylserine-specific lysophospholipase activity in humans. J. Biol. Chem. 1999, 274, 11053–11059.
- Hiramatsu, T.; Sonoda, H.; Takanezawa, Y.; Morikawa, R.; Ishida, M.; Kasahara, K.; Sanai, Y.R.; Taguchi, J.; Aoki, H.A. Biochemical and molecular characterization of two phosphatidic acid-selective phospholipase A1s, mPA-PLA1alpha and mPA-PLA1beta. J. Biol. Chem. 2003, 278, 49438–49447.
- Hide, W.A.; Chan, L.; Li, W.H. Structure and evolution of the lipase superfamily. J. Lipid. Res. 1992, 33, 167–178.
- Carriere, F.; Withers, M.C.T.H.; van Roussel, A.; Cambillau, C.; Verger, R. Structural basis for the substrate selectivity of pancreatic lipases and some related proteins. Biochim. Biophys. Acta 1998, 1376, 417–432.
- Winkler, F.K.; D’Arcy, A.; Hunziker, W. Structure of human pancreatic lipase. Nature 1990, 343, 771–774.
- Carriere, F.; Thirstrup, K.; Hjorth, S.; Ferrato, F.; Nielsen, P.F.; Withers-Martinez, C.; Cambillau, C.; Boel, E.; Thim, L.; Vergerc, R. Pancreatic lipase structure-function relationships by domain exchange. Biochemistry 1997, 36, 239–248.
- Higgs, H.N.; Glomset, J.A. Identification of a phosphatidic acid-preferring phospholipase A1 from bovine brain and testis. Proc. Natl. Acad. Sci. USA 1994, 91, 9574–9578.
- Higgs, H.N.; Han, M.H.; Johnson, G.E.; Glomset, J.A. Cloning of a phosphatidic acid-preferring phospholipase A1 from bovine testis. J. Biol. Chem. 1998, 273, 5468–5477.
- Nakajima, K.; Sonoda, H.; Mizoguchi, T.; Aoki, J.; Arai, H.; Nagahama, M.; Tagaya, M.; Tani, K. A novel phospholipase A1 with sequence homology to a mammalian Sec23p-interacting protein, p125. J. Biol. Chem. 2002, 277, 11329–11335.
- Araki, M.; Ohshima, N.; Aso, C.; Konishi, A.; Obinata, H.; Tatei, K.; Izumi, T. Enzymatic characterization of recombinant rat DDHD2: A soluble diacylglycerol lipase. J. Biochem. 2016, 160, 269–279.
- Inloes, J.M.; Hsu, K.L.; Dix, M.M.; Viader, A.; Masuda, K.; Takei, T.; Wood, M.R.; Cravatt, B.F. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc. Natl. Acad. Sci. USA 2014, 111, 14924–14929.
- Tani, K.; Mizoguchi, T.; Iwamatsu, A.; Hatsuzawa, K.; Tagaya, M. p125 is a novel mammalian Sec23p-interacting protein with structural similarity to phospholipid-modifying proteins. J. Biol. Chem. 1999, 274, 20505–20512.
- Quistad, G.B.; Barlow, C.; Winrow, C.J.; Sparks, S.E.; Casida, J.E. Evidence that mouse brain neuropathy target esterase is a lysophospholipase. Proc. Natl. Acad. Sci. USA 2003, 100, 7983–7987.
- Zaccheo, O.; Dinsdale, D.; Meacock, P.A.; Glynn, P. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J. Biol. Chem. 2004, 279, 24024–24033.
- Muhlig-Versen, M.; da Cruz, A.B.; Tschape, J.A.; Moser, M.; Buttner, R.; Athenstaedt, K.; Glynn, P.; Kretzschmar, D. Loss of Swiss cheese/neuropathy target esterase activity causes disruption of phosphatidylcholine homeostasis and neuronal and glial death in adult Drosophila. J. Neurosci. 2005, 25, 2865–2873.
- Kienesberger, P.C.; Lass, A.; Preiss-Landl, K.; Wolinski, H.; Kohlwein, S.D.; Zimmermann, R.; Zechner, R. Identification of an insulin-regulated lysophospholipase with homology to neuropathy target esterase. J. Biol. Chem. 2008, 283, 5908–5917.
- Liu, X.; Moon, S.H.; Jenkins, C.M.; Sims, H.F.; Gross, R.W. Cyclooxygenase-2 Mediated Oxidation of 2-Arachidonoyl-Lysophospholipids Identifies Unknown Lipid Signaling Pathways. Cell Chem. Biol. 2016, 23, 1217–1227.
- Mancuso, D.J.; Sims, H.F.; Han, X.; Jenkins, C.M.; Guan, S.P.; Yang, K.; Moon, S.H.; Pietka, T.; Abumrad, N.A.; Schlesinger, P.H.; et al. Genetic ablation of calcium-independent phospholipase A2gamma leads to alterations in mitochondrial lipid metabolism and function resulting in a deficient mitochondrial bioenergetic phenotype. J. Biol. Chem. 2007, 282, 34611–34622.
- Pickard, R.T.; Strifler, B.A.; Kramer, R.M.; Sharp, J.D. Molecular cloning of two new human paralogs of 85-kDa cytosolic phospholipase A2. J. Biol. Chem. 1999, 274, 8823–8831.
- Song, C.; Chang, X.J.; Bean, K.M.; Proia, M.S.; Knopf, J.L.; Kriz, R.W. Molecular characterization of cytosolic phospholipase A2-beta. J. Biol. Chem. 1999, 274, 17063–17067.
- Uyama, T.; Morishita, J.; Jin, X.H.; Okamoto, Y.; Tsuboi, K.; Ueda, N. The tumor suppressor gene H-Rev107 functions as a novel Ca2+-independent cytosolic phospholipase A1/2 of the thiol hydrolase type. J. Lipid. Res. 2009, 50, 685–693.
- Robinson, M.; Waite, M. Physical-chemical requirements for the catalysis of substrates by lysosomal phospholipase A1. J. Biol. Chem. 1983, 258, 14371–14378.
- Kucera, G.L.; Miller, C.; Sisson, P.J.; Wilcox, R.W.; Wiemer, Z.; Waite, M. Hydrolysis of thioester analogs by rat liver phospholipase A1. J. Biol. Chem. 1988, 263, 12964–12969.
- Ueda, H.; Kobayashi, T.; Kishimoto, M.; Tsutsumi, T.; Okuyama, H. A possible pathway of phosphoinositide metabolism through EDTA-insensitive phospholipase A1 followed by lysophosphoinositide-specific phospholipase C in rat brain. J. Neurochem. 1993, 61, 1874–1881.
- Ueda, H.; Kobayashi, T.; Kishimoto, M.; Tsutsumi, T.; Watanabe, S.; Okuyama, H. The presence of Ca(2+)-independent phospholipase A1 highly specific for phosphatidylinositol in bovine brain. Biochem. Biophys. Res. Commun. 1993, 195, 1272–1279.
- Ueda, H.; Kobayashi, T.; Kishimoto, M.; Tsutsumi, T.; Okuyama, H. EDTA-insensitive deacylation of phosphatidylinositol in porcine platelet membranes. Life Sci. 1993, 53, 629–634.
- Hostetler, K.Y.; Gardner, M.F.; Giordano, J.R. Purification of lysosomal phospholipase A and demonstration of proteins that inhibit phospholipase A in a lysosomal fraction from rat kidney cortex. Biochemistry 1986, 25, 6456–6461.
- Waite, M. The Phospholipases: Handbook of Lipid Research; Plenum Publishing Corp: New York, NY, USA, 1987; pp. 79–110.
- Van den Bosch, H. Phosphaolipases; Elsevier: Amsterdam, The Netherlands, 1982; pp. 313–351.
- Pete, M.J.; Ross, A.H.; Exton, J.H. Purification and properties of phospholipase A1 from bovine brain. J. Biol. Chem. 1994, 269, 19494–19500.
- Uchiyama, S.; Miyazaki, Y.; Amakasu, Y.; Kuwata, H.; Nakatani, Y.; Atsumi, G.; Murakami, M.; Kudo, I. Characterization of heparin low-affinity phospholipase A1 present in brain and testicular tissue. J. Biochem. 1999, 125, 1001–1010.
- Horigome, K.; Hayakawa, M.; Inoue, K.; Nojima, S. Selective release of phospholipase A2 and lysophosphatidylserine-specific lysophospholipase from rat platelets. J. Biochem. 1987, 101, 53–61.
- Horigome, K.; Hayakawa, M.; Inoue, K.; Nojima, S. Purification and characterization of phospholipase A2 released from rat platelets. J. Biochem. 1987, 101, 625–631.
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
3.3K
Revisions:
3 times
(View History)
Update Date:
27 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





