Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tariq Pervaiz | + 3119 word(s) | 3119 | 2021-12-29 04:00:52 | | | |
| 2 | Jessie Wu | Meta information modification | 3119 | 2022-01-13 02:15:10 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Pervaiz, T. MicroRNA and cDNA-Microarray against Abiotic Stress Response. Encyclopedia. Available online: https://encyclopedia.pub/entry/18130 (accessed on 07 February 2026).
Pervaiz T. MicroRNA and cDNA-Microarray against Abiotic Stress Response. Encyclopedia. Available at: https://encyclopedia.pub/entry/18130. Accessed February 07, 2026.
Pervaiz, Tariq. "MicroRNA and cDNA-Microarray against Abiotic Stress Response" Encyclopedia, https://encyclopedia.pub/entry/18130 (accessed February 07, 2026).
Pervaiz, T. (2022, January 12). MicroRNA and cDNA-Microarray against Abiotic Stress Response. In Encyclopedia. https://encyclopedia.pub/entry/18130
Pervaiz, Tariq. "MicroRNA and cDNA-Microarray against Abiotic Stress Response." Encyclopedia. Web. 12 January, 2022.
Copy Citation
The miRNAs and cDNA-microarrays are powerful tools to enhance abiotic stress tolerance in plants through multiple advanced sequencing and bioinformatics techniques, including miRNA-regulated network, miRNA target prediction, miRNA identification, expression profile, features (disease or stress, biomarkers) association, tools based on machine learning algorithms, NGS, and tools specific for plants. Such technologies were established to identify miRNA and their target gene network prediction, emphasizing current achievements, impediments, and future perspectives.
salinity stress
cold stress
miRNAs
1. miRNAs and cDNA-Microarray Associated with Cold Stress
Cold stress (frost and chilling) decreases crop yields worldwide through tissue degradation and delayed growth. Most temperate plants have evolved cold resistance through cold-acclimatization [1]. Signaling pathways were being used in response to winter stress. The functional genes transform reactions, and reposts suggest that the signaling pathways for leaf senescence and plant defense responses may overlap [2]. The most characteristic region of cold-stress responsive genes includes transcription factors, such as CBF/DREB and stress-inducible candidate genes, identified as KIN (cold-induced), COR (cold-regulated), and LTI genes (induced by low temperature) or RD (dehydration) [3]. Several HSPs (heat shock proteins) are also reported for their functions against cold stress. HSPs, which perform as molecular chaperons, play an important regulatory function in protecting from stress by restoring normal protein conformation and thus maintaining cellular homeostasis in plants [4]. The number of the miRNA target genes in expression is intricate during stress and plant growth. These miRNAs are co-regulated by both developmental signals and ecological factors (Table 3). The cold-responsive miRNAs were detected by microarray analysis in Arabidopsis thaliana (miR165, miR31, miR156, miR168, miR171, miR396) and recommended by identifying their expression patterns in their promoter sequences and evaluating the cis-components (Table 3, Figure 1) [5][6]. Furthermore, high-intensity light (HL) responsive genes were assessed with the drought-inducible genes reported with a similar microarray system, which exposed an impenetrable intersection between drought and HL-induced genes. Moreover, 10 genes were identified as being involved in the regulation by HL, drought, salinity, and cold stress (Table 1 and Table 2). These genes are comprised of ERD10, RD29A, KIN1, LEA14, COR15a, and ERD7, and most of them are considered to be concerned in the defense of cellular components [7][8][9]. Along with the HL-inducible genes, some are also identified and encouraged by other stresses (heat, drought, and cold), including AtGolS, LEA, RAB, RD, COR, ERD, HSP, KIN, lipid-transfer proteins, and fibrillins [10][11][12].
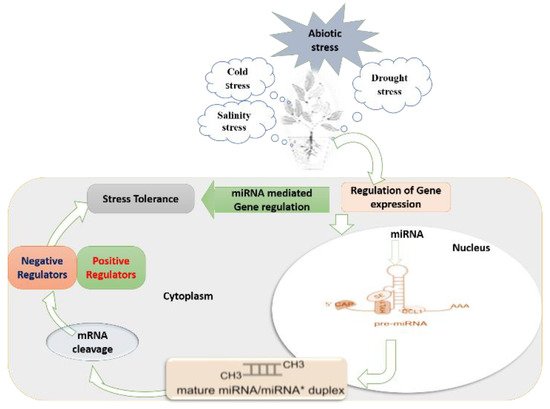
Figure 1. Schematic summary of miRNA-mediated regulatory mechanisms under abiotic stress in plant cells, with the particular formation process of miRNAs and miRNA mediated gene regulation: (1) miRNA gene is transcribed to a long sequence of primary miRNA (pri-miRNA). Primary miRNAs (pri-miRNAs) are transcribed from nuclear-encoded MIR genes by RNA polymerase II (Pol II), leading to precursor transcripts with a characteristic hairpin structure. (2) The pri-miRNA is cleaved to a stem-loop intermediate called miRNA precursor or pre-miRNA.
Table 1. Examples of miRNAs identified in model plants under drought, cold and salinity stresses.
| Stress Condition | Plant Species | Inducible Genes | Known Responsive miRNAs | Functions | References |
|---|---|---|---|---|---|
| Drought stress | Arabidopsis thaliana | Rd29A (At5g52310) CCAAT-binding transcription factors |
miR164, miR169, miR389, miR393, miR396, miR397, miR402 |
Pathogen immune response Drought tolerance Oxidative stress tolerance Pathogen immunity response Syncytium formation response to parasitic nematodes |
[13][14][15][16] |
| Medicago truncatula | CCAAT Binding Factor (CBF) Growth Regulating Factor (GRF) Cu/Zn superoxide dismutases (CSD1, CSD2) TIR-NBS-LRR domain protein |
miR169, miR396 miR398, miR2118 |
Drought tolerance Syncytium formation response to parasitic nematodes Oxidative stress tolerance Photoperiod-sensitive male sterility |
[13][17] | |
| Oryza sativa | SalT (LOC_Os01g24710) TIR1 OsLEA3 (LOC_Os05g46480) |
miR393 miR402 |
Salt/cold tolerance | [18][14][15][19] | |
| Cold stress | Arabidopsis thaliana | Rd29A (At5g52310) CBF3 (At4g25480) |
miR165, miR172, miR169, miR396, miR397, miR402 |
Drought/cold tolerance Drought tolerance Heat stress tolerance |
[13][14] |
| Oryza Sativa | OsWRKY71 (LOC_Os02g08440) OsMAPK2(LOC_Os03g17700) Os05g47550, Os03g42280 Os01g73250, Os12g16350 Os03g19380 |
miR319, miR389, miR393, miR1320, miR1435 miR1884b, CHY1 CP12-2 |
Drought/salt tolerance Cold tolerance Pathogen immunity response |
[14][19][20][21] | |
| Salinity stress | Arabidopsis thaliana | Rd29A (At5g52310) COR15A (At2g42540) |
miR389, miR393, | Oxidative stress tolerance Heat stress tolerance |
[22] |
| Populus trichocarpa | Dihydropyrimidinase | miR162, miR164, miR166, miR167, miR168, miR172, miR395, miR396 | Pathogen immune response Drought tolerance Drought/cold tolerance Sulfate-deficiency response |
[23][24][25] | |
| Glycine max | miR1507a, miR395 | Sulfate-deficiency response | [26] | ||
| Oryza sativa | SalT (LOC_Os01g24710) OsLEA3 (LOC_Os05g46480) |
miR156, miR158, miR159, miR397, miR398, miR482.2, miR530a, miR1445 | Drought tolerance Pathogen immune response Heat stress tolerance |
[20][27][28][29] | |
| Zea mays | miR402 | Seed germination and seedling growth of Arabidopsis under stress | [15] |
Table 2. Microarray analysis of genes involved in the drought, salinity and cold stress responses in Arabidopsis.
| Phenotype of Mutants | Genes | Function | AGI Code | Coded Proteins | Microarrays | |
|---|---|---|---|---|---|---|
| Increased tolerance to drought |
AtPARP2 | DNA repair | At2g31320 | Poly (ADPribose) polymerase | 24K Affymetrix |
[30][31][32] |
| Hypersensitive to drought stress |
AHK1/ ATHK1 |
positive regulator of drought and salt stress responses | At2g17820 | Histidine kinase | 22K Agilent | [30][33][34] |
| Increased tolerance to drought stress |
AREB1/ ABF2 |
regulate the ABRE-dependent expression | At1g45249 | bZIP TF | 22K Agilent | [31][35][36] |
| Increased tolerance to salt stress |
AtbZIP60 | encodes a predicted protein of 295 aa | At1g42990 | bZIP TF | 44K Agilent | [35][37] |
| Increased tolerance to drought stress |
AtMYB60 | regulates stomatal movements and plant drought tolerance | At1g08810 | MYB TF | 7K cDNA | [38] |
| Increased sensitivity to drought stress |
AtMYB41 | control of primary metabolism and negative regulation | At4g28110 | MYB TF | 24K Affymetrix |
[39][40] |
| Increased tolerance to drought and salt stress |
AHK2 | positive regulators for cytokinin signaling | At5g35750 | Histidine kinase | Agilent | [33][34] |
| Increased tolerance to drought and salt stress |
AHK3 | perception of cytokinin, downstream signal transduction | At1g27320 | Histidine kinase | 22K Agilent | [33][34] |
| Increased tolerance to drought and freezing stress |
DREB1A/ CBF3 |
stress-inducible transcription factor | ERF/AP2 TF | ERF/AP2 TF | 1.3K cDNA | [41] |
| Increased tolerance to drought stress |
DREB2A | heat shock-stress responses. | At5g05410 | ERF/AP2 TF | 22K Agilent 7K cDNA |
[42] |
| Hypersensitive to salt |
HOS10 | coordinating factor for responses to abiotic stress and for growth and development. | At1g35515 | MYB TF | 24K Affymetrix |
[30][43] |
| Increased tolerance to drought stress |
ZFHD1 | mediates all the protein-protein interactions | At1g69600 | Zinc finger HD TF |
22K Agilent | [34][37] |
Table 3. miRNAs regulated by drought stress, salinity stress, and cold stress in plants.
| Stress Condition | Plant Species | miRNA | Key Functions | Response | References |
|---|---|---|---|---|---|
| Drought stress | Medicago truncatula | miR398a,b miR408 miR399k miR2089 miR2111a-f,h-s miR2111g miR4414a |
Oxidative stress tolerance Salt/drought/cold/oxidative osmotic-stress responses Phosphate-deficiency response |
Up-regulated | [20][44][45][46][47] |
| miR398b,c miR2111u,v miR5274b miR1510a-3p, 5p miR1510a |
Heat stress tolerance Drought responsive Oxidative-stress tolerance triggering phasiRNA production from numerous NB-LRRs |
Down-regulated | [44][46][47] | ||
| Glycine max | miR5554a-c | Drought responsive | [46] | ||
| Salinity stress | Glycine max | miR169d miR395a miR395b,c miR1510a-5p miR1520d,e,l,n,q |
Drought tolerance Sulfate-deficiency response triggering phasiRNA production from numerous NB-LRRs |
Up-regulated | [20][48][49] |
| gma-miR159b,c gma-miR169b,c gma-miR1520c |
Pathogen immune response Drought/Salt tolerance |
Down-regulated | [49] | ||
| Phaseolus vulgaris | pvu-miR159.2 | Plant–nematode interaction | [31] | ||
| Cold stress | Phaseolus vulgaris | pvu-miR2118 | regulate the expression of genes encoding the TIR-NBS-LRR resistance protein | Up-regulated |
DNA microarrays almost in all genes of the unicellular Synechocystis sp PCC6803 were used to investigate the gene expression sequential software [50]. A cDNA-microarray was used to test the profile expression in cold stress, and 328 temperature-regulated transcripts were reported. OsMYB3R-2 was studied further and was shown to be a dominant regulator against stress [51]. In this study, there was an attempt to use a 3.1K cDNA-microarray to express the cold-regulated transcripts in the Capsicum annuum. Several TFs, including the EREBP (CaEREBP-C1 to C4) family of four genes, a protein of the ring domain, a bZIP protein (CaBZ1), RVA1, a WRKY (CaWRKY1), and HSF1 protein have been observed among the cold stress-regulated genes. These genes included CaBZ1, CaEREBP-C3, NtPRp27, the SAR8.2 protein precursor, putative trans-activator factor, malate hydrogenase, putative protein of auxin-repressed, xyloglu-canendo-1, 4-D-gucanase precursor, LEA protein 5 (LEA5), homologous DNAJ protein, PR10 and Stns LTP [52][53]. cDNA microarray z1300 full-length cDNAs were used in Arabidopsis to identify cold stress-inducing genes and target genes of DREB1A/CBF3. Six genes were documented based on microarray and, in RNA gel blot analyses, it was observed that a novel DREB1A controls cold- and drought-inducible genes [41][54]. Furthermore, microarray with full-length cDNA was performed by 1300 full-length cDNAs and cDNA microarray to discover cold-induced genes. Previous reposts exhibited the target genes of DREB1A/CBF3 and stress-inducible gene expressions were controlled by transcription factors [10]; in contrast, stress-sensitive genes’ expressions were reported as specific to the growth stage [40]. Full-length cDNA microarray is convenient for analyzing the Arabidopsis gene expression patterns under cold stress, and can also be used to identify the functional genes of stress-related TFs that are likely to act as DNA elements by merging the genomic sequence data with the expression data [10][55]. Additionally, cold stress is also induced by the increase in the proline content in plants (osmoprotectant). Microarray and RNA gel blot research found that the proline can induce the expression of several genes with the proline-responsive elements in their promoters (PRE, ACTCAT) [11][55][56]. Microarray analysis was carried out to detect the cold-inducible AP2 gene family transcription factor RAV1 [57], which could control plant growth under stress. RAV1 is down-regulated by epibrassinolide, and transgenic Arabidopsis overexpressing RAV1 exhibits a rosette leaf and adjacent root growth retardation, although the early-flowering phenotype showed antisense to RAV1 plants [58][59].
2. miRNAs and cDNA-Microarray Response to Salinity Stress
Salt intrusion from saline soils and irrigation water is one of the most severe and harmful risks to reduce agricultural production and adverse effects on cultivated land and the geographical distribution of plant species [60][61][62], coupled with oxidative stress [63]. The most imperative cations in saline soils are calcium, potassium, magnesium, and sodium, and the main anions in saline soils are chloride, bicarbonate, sulfate, nitrate, and carbonates. Other electrolytes causative to salinity are borane, molybdenum, strontium, silicon dioxide, aluminum cation, and barium ion [64][65]. Higher concentrations of sodium chloride (NaCl) typically affect plant development, metabolism, and physiology at various metabolic phases (ion toxicity, nutrient imbalance, and oxidative stress) [60][66]. Despite such advances in scientific research, it remains unclear about the underlying molecular mechanism of salinity responses in plants. However, based on the combination of microarray and inhibition subtractive hybridization (SSH), changes in the transcriptome profile caused by salt induction were studied and evaluated [67]. Investigation of complete transcriptomics suggests that these processes, such as the synthesis of osmolytes and ion carriers and the regulation of transcription and translation mechanisms, have distinctive reactions under salinity stress. In particular, the introduction of transcripts of specific TFs, ribosomal genes, RNA-binding proteins, and translation initiation and elongation factors has been testified [68][69].
Using cDNA microarray in Synechocystis, 19 genes were reported to be instantaneously regulated under salinity stress. The salt- and osmo-regulated genes, and some putative sensor molecules, have been implicated during salinity stress signaling [33]. Several differentially regulated miRNAs have been reported against salinity stress. In A. thaliana, several microRNAs are regulated against salinity stress, such as miR156, miR158, miR159, miR165, miR167, miR168, miR169, miR171, miR319, miR393, miR394, miR396, and miR397 (Table 3, Figure 2) [70]. In Populus trichocarpa, miR1445, miR1447, miR1446a-e, miR530a, and miR171l-n were down-regulated (Table 3) [71]. Arenas-Huertero et al. [29] reported, in Proteus vulgaris, the production of miRS1 and miR159.2 expression in response to salinity. Furthermore, miR169g and family members of miR169n were induced in saline-rich conditions [72]. However, there is a need to discover and annotate novel functional genes which have a probable function against salinity stress. Subsequently, a large number of genes in plants still have unknown functions [73]. Recent studies revealed that specific down-regulation of the bacterial-type phosphoenolpyruvate carboxylase (PEPC) gene Atppc4 by artificial microRNA enhanced the salinity tolerance in A. thaliana. The increased salinity tolerance might be linked to enhanced PEPC activity [74][75]. Transcript control for salinity-tolerant rice with microarrays, like 1728 cDNAs from salinity-stressed roots libraries, was studied in response to high salinity (Table 3) [75][76][77].
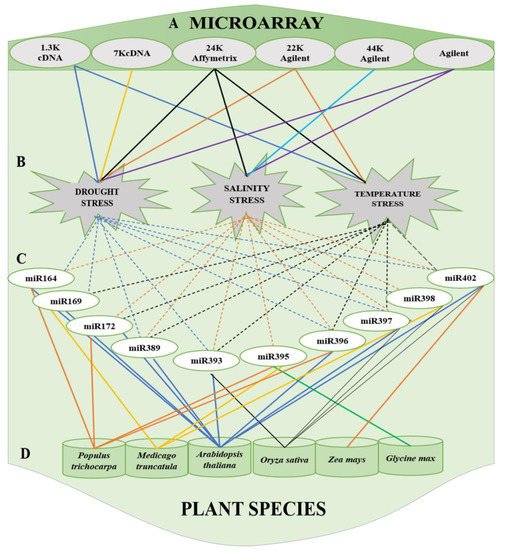
Figure 2. Summary of commonly used (A) microarrays (cDNA, Affymetrix, and Agilent) to stress and (B) miRNAs, categorized based on the stress, that respond to drought stress, salinity and temperature stress and (C) miRNAs reported in (D) plant species: Populus trichocarpa, Medicago truncatula, Arabidopsis thaliana, Oryza sativa, Zea mays and Glycine max.
A tiling path microarray was used to examine the high-throughput expression profiling patterns under various environmental stresses for all of the known miRNAs [13][60] (Table 1 and Table 4). The analysis revealed that the effects of miRNAs under low-temperature, drought, and high salinity with miRNA chips represent, approximately, all of the reported miRNAs cloned or recognized in A. thaliana (L.). High salinity stress agitates homeostasis in water potential. Extreme changes in water homeostasis and ions lead to molecular breakdown, stunted growth, and even the death of cells or whole plants [13][44].
Table 4. Software and tools used for the detection of plant miRNA and cDNA microarray data analysis.
| Software and Tools | Function | Website | Reference | Accessed |
|---|---|---|---|---|
| Software and tools used for detection of plant miRNA and data analysis | ||||
| MiPred | Random forest (RF)-based miRNA predictor, which can distinguish between real and pseudo-miRNA precursors | http://server.malab.cn/MiPred/ | [45] | 5 November 2021 |
| miBridge | Algorithm and database | http://sitemaker.umich.edu/mibridge/home | [46] | 5 November 2021 |
| miRTar | A novel rule-based model learning method for cell line specific microRNA target prediction | http://miRTar.mbc.nctu.edu.tw | [45] | 5 November 2021 |
| PolymiRTS | Linking polymorphisms in microRNAs and their target sites | http://compbio.uthsc.edu/miRSNP | [47] | 25 November 2021 |
| miRGator | microRNA portal for deep sequencing, expression profiling and mRNA targeting | http://mirgator.kobic.re.kr | [48] | 10 November 2021 |
| Bowtie | Aligns efficiently, and short-read aligners | http://bowtie-bio.sourceforge.net | [45] | 5 November 2021 |
| miRBase | Provides handy and useful ID conversion tools | http://www.mirbase.org/ | [45] | 25 November 2021 |
| miRDB | miRNA target databases | http://www.mirdb.org | [49] | 25 November 2021 |
| mirDIP | Integrative database of microRNA target predictions | http://ophid.utoronto.ca/mirDIP | [78] | 25 November 2021 |
| miRanda | Predict or collect miRNA targets | http://34.236.212.39/microrna/home.do | [45] | 25 November 2021 |
| RNAhybrid | microRNA target prediction | https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid | [45] | 8 November 2021 |
| miTALOS | Analyzes tissue specific microRNA function. | http://mips.helmholtz-muenchen.de/mitalos | [79] | 5 November 2021 |
| RNA22 | microRNA target predictions | https://cm.jefferson.edu/rna22 | [80] | 5 November 2021 |
| psRNATarget | Small RNA target analysis server | http://plantgrn.noble.org/psRNATarget/ | [81] | 5 November 2021 |
| miRandola | Curated knowledge base of non-invasive biomarkers | http://mirandola.iit.cnr.it/ | [81] | 5 November 2021 |
| ChIPBase | Decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data | http://rna.sysu.edu.cn/chipbase/ | [81][82] | 1 October 2021 |
| MirGeneDB | Curated miRNA gene database | http://mirgenedb.org/ | [83] | 28 November 2021 |
| TarHunter | Predicting conserved microRNA targets and target mimics in plants | http://tarhunter.genetics.ac.cn | [84] | 28 November 2021 |
| TissueAtlas | Tissue specificity miRNA database | https://ccb-web.cs.uni-saarland.de/tissueatlas/ | [45] | 28 November 2021 |
| miRNAme Converter | miRNA ID converter | http://163.172.134.150/miRNAmeConverter-shiny | [85] | 28 November 2021 |
| Software and tools used for detection of plant microarray and data analysis | ||||
| Array Designer | Design primers and probes for oligo and cDNA expression microarrays. | http://www.premierbiosoft.com/dnamicroarray/index.html | [86] | 1 November 2021 |
| Stanford Microarray Database SMD | Stores raw and normalized data from microarray experiments | http://smd-www.stanford.edu//download/ | [87] | 1 November 2021 |
| eArray | Designing Agilent arrays | http://earray.chem.agilent.com/earray/login.do | [86] | 1 November 2021 |
| Significance Analysis of Microarrays | Adjustments for multiple testing, statistical analysis for discrete, quantitative, and time series data, gene set enrichment analysis | http://www-stat.stanford.edu/~tibs/SAM/ | [88] | 5 November 2021 |
| Visual OMP | Design software for RNA, DNA, single or multiple probe design, microarrays, Taq Manassays, genotyping, single and multiplex PCR, secondary structure simulation, sequencing, genotyping. | http://www.dnasoftware.com/Products/VisualOMP | [86] | 5 November 2021 |
| caArray | Open-source, web and programmatically accessible microarray data management system that supports the annotation of microarray | http://caarray.nci.nih.gov/ | 5 November 2021 | |
| Gene Expression Model Selector | Diagnostic models and biomarker discovery | http://www.gems-system.org/ | [89] | 18 November 2021 |
| Gene index | Gene Index Project is to use the available EST and gene sequences, along with the reference genomes, to provide an inventory of likely genes and variants. | http://compbio.dfci.harvard.edu/tgi/plant.html | [86] | 5 November 2021 |
| Genesis | Java package of tools to simultaneously visualize and analyze a whole set of gene expression experiments | http://genome.tugraz.at/genesisclient/genesisclient_description.shtml | 18 November 2021 | |
| RMA Express | Standalone GUI program for Windows, OS X and Linux to compute gene expression summary values for Affymetrix | http://rmaexpress.bmbolstad.com http://www.r-project.org http://www.bioconductor.org |
18 November 2021 | |
| dCHIP | Model-based expression analysis for Affymetrix gene expression arrays | http://www.dchip.org | [90] | 18 November 2021 |
| TM4 | Microarray Data Manager (MADAM), TIGR Spotfinder, Microarray Data Analysis System (MIDAS), and Multi experiment Viewer (MeV) | http://www.tm4.org/ | [90] | 18 November 2021 |
| Able Image Analyser | Software for image analysis. It enables dimensional measurements: distance, area, angle in digital images | http://able.mulabs.com | [86] | 18 November 2021 |
| ImaGene | Unique, robust, room-temperature preservation solutions for nucleic acids, biospecimens and bioreagents for in the living ectors | http://www.biodiscovery.com/index/imagene | [86] | 13 November 2021 |
| Spotfinder | Custom-designed cDNA array, the chips are scanned using a microarray scanner | http://www.tm4.org/spotfinder.html | [90] | 18 November 2021 |
| SNOMAD | Web-based tool and has various normalization options for two-channel and single-channel experiments | http://pevsnerlab.kennedykrieger.org/snomadinput.html | [90] | 18 November 2021 |
| Multiexperimet Viewer | Cloud-based application supporting analysis, visualization, and stratification of large genomic data | http://www.tm4.org/mev.html | 18 November 2021 | |
| Onto-Express and Pathway-Express | Automatically translates DE gene transcripts from microarray experiments into functional profiles characterizing the impact of the condition studied | http://vortex.cs.wayne.edu/projects.htm | [90] | 13 November 2021 |
| DAVID/EASE | Database for annotation, visualization and integrated discovery (DAVID) is an online tool for annotation and functional analysis. Expression analysis systematic sxplorer (EASE) | http://david.abcc.ncifcrf.gov | [90] | 13 November 2021 |
Oligo-DNA microarrays were developed in common wheat, and these microarrays were designed to include approximately 32,000 distinctive genes characterized by several expressed sequence tags (ESTs). To classify the salinity-stress responsive genes, the expression profiles of transcripts that responded to stress were examined using microarrays. It was concluded that 5996 genes were verified by more than a 2-fold change in expression. These genes were categorized into twelve groups based on gene expression patterns [91]. Transcription-regulator activity, DNA binding, and the genes’ assigned transcription factor functions were preferentially classified as immediate response genes. In wheat, candidate genes were identified as involved in salinity-stress tolerance [91][92]. These genes are active in the regulation of transcription [1][73] and the signal transduction that is engaged in metabolic pathways [93] or acting as ion transporters [94]. cDNA library in yeast (Saccharomyces cerevisiae) was examined using a synthetic medium augmented with excessive salt concentrations (900 mM). A few clones showed comparatively improved growth. The notorious clones bore the Guanyl transferase (OsMPG1) mannose-1-phosphate gene [62]. Extreme salinity stress was significantly linked with the transcription factors of four tomato genes from the family of zinc finger. There has been prior evidence of the relationship between zinc finger transcription factors and plant salinity tolerance [95][96]. Overexpression of OSISAP1 in transgenic tobacco resulted in tolerance to salinity, dehydration, and cold stress in the new sprouts [97].
A microarray containing 384 genes associated with stress responses was used in Medicago truncatula genotypes (Jemalong A17 and 108-R) to compare rooting gene expression during salt stress. The homolog of flora TFIIIA-related TF, MtZpt2-1, and COLD-REGULATEDA1 genes were known to regulate the previous genes and were acknowledged in Jemalong A17 stress-tolerant genotypes. Two MtZpt2 Transcription factors (MtZpt2-1 and MtZpt2-2) have shown increased expression in the roots compared to 108-R [98]. Salinity stress is attributed to diverse stresses that persuade overlapping patterns in gene expression. For example, in an investigation of 8100 A. thaliana genes, approximately 2400 genes were reported to have a widespread expression in exposure to salt, oxidative and cold stress [99]. In addition, 23 genes were reported against NaCl stress. This also accounted for a small percentage of DEGs, including encoding transcription factors WOX2 and BZIP3, calcium-binding protein CML42, ubiquitin-protein ligase UBC17, and IDA-like 5 protein [99]. Most prominently, synthesized isiA encoded a novel chlorophyll (Chl)-binding protein [100] (Table 3).
References
- Sunkar, R.; Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12, 301–309.
- Trindade, I.; Capitão, C.; Dalmay, T.; Fevereiro, M.P.; Dos Santos, D.M. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2010, 231, 705–716.
- Fang, Z.; Zhang, X.; Gao, J.; Wang, P.; Xu, X.; Liu, Z.; Shen, S.; Feng, B. A buckwheat (Fagopyrum esculentum) DRE-binding transcription factor gene, FeDREB1, enhances freezing and drought tolerance of transgenic Arabidopsis. Plant Mol. Biol. Report. 2015, 33, 1510–1525.
- Wang, T.; Chen, L.; Zhao, M.; Tian, Q.; Zhang, W.-H. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genom. 2011, 12, 1–11.
- Mantri, N.; Basker, N.; Ford, R.; Pang, E.; Pardeshi, V. The role of micro-ribonucleic acids in legumes with a focus on abiotic stress response. Plant Genome 2013, 6.
- Li, H.; Dong, Y.; Yin, H.; Wang, N.; Yang, J.; Liu, X.; Wang, Y.; Wu, J.; Li, X. Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biol. 2011, 11, 1–12.
- Dong, Z.; Shi, L.; Wang, Y.; Chen, L.; Cai, Z.; Wang, Y.; Jin, J.; Li, X. Identification and dynamic regulation of microRNAs involved in salt stress responses in functional soybean nodules by high-throughput sequencing. Int. J. Mol. Sci. 2013, 14, 2717–2738.
- Arenas-Huertero, C.; Pérez, B.; Rabanal, F.; Blanco-Melo, D.; De la Rosa, C.; Estrada-Navarrete, G.; Sanchez, F.; Covarrubias, A.A.; Reyes, J.L. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol. Biol. 2009, 70, 385–401.
- Albert, E. Genetic and Genomic Determinants of Response to Water Deficit in Tomato (Solanum lycopersicum) and Impact on Fruit Quality. Ph.D. Thesis, Université d’Avignon, Avignon, France, 2017.
- Seki, M.; Narusaka, M.; Kamiya, A.; Ishida, J.; Satou, M.; Sakurai, T.; Nakajima, M.; Enju, A.; Akiyama, K.; Oono, Y. Functional annotation of a full-length Arabidopsis cDNA collection. Science 2002, 296, 141–145.
- Satoh, R.; Nakashima, K.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ACTCAT, a novel cis-acting element for proline-and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis. Plant Physiol. 2002, 130, 709–719.
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016, 7, 114.
- Zhao, B.; Liang, R.; Ge, L.; Li, W.; Xiao, H.; Lin, H.; Ruan, K.; Jin, Y. Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 2007, 354, 585–590.
- Sunkar, R.; Zhu, J.-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019.
- Wang, Y.; Long, L.-H. Identification and isolation of the coldresistance related miRNAs in Pisum sativum Linn. J. Liaoning Norm. Univ. 2010, 2, 027.
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726.
- Sunkar, R.; Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12, 301–309.
- Meng, Y.; Ma, X.; Chen, D.; Wu, P.; Chen, M. MicroRNA-mediated signaling involved in plant root development. Biochem. Biophys. Res. Commun. 2010, 393, 345–349.
- Raffaele, S.; Mongrand, S.; Gamas, P.; Niebel, A.; Ott, T. Genome-wide annotation of remorins, a plant-specific protein family: Evolutionary and functional perspectives. Plant Physiol. 2007, 145, 593–600.
- Zhou, X.; Jeker, L.T.; Fife, B.T.; Zhu, S.; Anderson, M.S.; McManus, M.T.; Bluestone, J.A. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J. Exp. Med. 2008, 205, 1983–1991.
- Rabbani, M.A.; Maruyama, K.; Abe, H.; Khan, M.A.; Katsura, K.; Ito, Y.; Yoshiwara, K.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003, 133, 1755–1767.
- Moons, A.; Prinsen, E.; Bauw, G.; Van Montagu, M. Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell 1997, 9, 2243–2259.
- Boualem, A.; Laporte, P.; Jovanovic, M.; Laffont, C.; Plet, J.; Combier, J.P.; Niebel, A.; Crespi, M.; Frugier, F. MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. 2008, 54, 876–887.
- Yang, F.; Yu, D. Overexpression of Arabidopsis MiR396 enhances drought tolerance in transgenic tobacco plants. Acta Bot. Yunnanica 2009, 31, 421–426.
- Subramanian, S.; Fu, Y.; Sunkar, R.; Barbazuk, W.B.; Zhu, J.-K.; Yu, O. Novel and nodulation-regulated microRNAs in soybean roots. BMC Genom. 2008, 9, 1–14.
- Zeng, H.Q.; Zhu, Y.Y.; Huang, S.Q.; Yang, Z.M. Analysis of phosphorus-deficient responsive miRNAs and cis-elements from soybean (Glycine max L.). J. Plant Physiol. 2010, 167, 1289–1297.
- Cui, H.; Zhai, J.; Ma, C. miRLocator: Machine learning-based prediction of mature microRNAs within plant pre-miRNA sequences. PLoS ONE 2015, 10, e0142753.
- Yu, Y.; Wu, G.; Yuan, H.; Cheng, L.; Zhao, D.; Huang, W.; Zhang, S.; Zhang, L.; Chen, H.; Zhang, J.; et al. Identification and characterization of miRNAs and targets in flax (Linum usitatissimum) under saline, alkaline, and saline-alkaline stresses. BMC Plant Biol. 2016, 16, 124.
- Arenas-Huertero, C.; Pérez, B.; Rabanal, F.; Blanco-Melo, D.; De la Rosa, C.; Estrada-Navarrete, G.; Sanchez, F.; Covarrubias, A.A.; Reyes, J.L. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol. Biol. 2009, 70, 385–401.
- Gu, Z.; Pan, W.; Chen, W.; Lian, Q.; Wu, Q.; Lv, Z.; Cheng, X.; Ge, X. New perspectives on the plant PARP family: Arabidopsis PARP3 is inactive, and PARP1 exhibits predominant poly (ADP-ribose) polymerase activity in response to DNA damage. BMC Plant Biol. 2019, 19, 364.
- Rasheed, S.; Bashir, K.; Matsui, A.; Tanaka, M.; Seki, M. Transcriptomic Analysis of Soil-Grown Arabidopsis thaliana Roots and Shoots in Response to a Drought Stress. Front. Plant Sci. 2016, 7, 180.
- Vanderauwera, S.; De Block, M.; Van de Steene, N.; Van De Cotte, B.; Metzlaff, M.; Van Breusegem, F. Silencing of poly (ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc. Natl. Acad. Sci. USA 2007, 104, 15150–15155.
- Marin, K.; Suzuki, I.; Yamaguchi, K.; Ribbeck, K.; Yamamoto, H.; Kanesaki, Y.; Hagemann, M.; Murata, N. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 2003, 100, 9061–9066.
- Tran, L.-S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628.
- Seki, M.; Narusaka, M.; Ishida, J.; Nanjo, T.; Fujita, M.; Oono, Y.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002, 31, 279–292.
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488.
- Fujita, M.; Mizukado, S.; Fujita, Y.; Ichikawa, T.; Nakazawa, M.; Seki, M.; Matsui, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Identification of stress-tolerance-related transcription-factor genes via mini-scale Full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem. Biophys. Res. Commun. 2007, 364, 250–257.
- Cominelli, E.; Galbiati, M.; Vavasseur, A.; Conti, L.; Sala, T.; Vuylsteke, M.; Leonhardt, N.; Dellaporta, S.L.; Tonelli, C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 2005, 15, 1196–1200.
- Wang, R.; Okamoto, M.; Xing, X.; Crawford, N.M. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003, 132, 556–567.
- Padmalatha, K.V.; Dhandapani, G.; Kanakachari, M.; Kumar, S.; Dass, A.; Patil, D.P.; Rajamani, V.; Kumar, K.; Pathak, R.; Rawat, B. Genome-wide transcriptomic analysis of cotton under drought stress reveal significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes. Plant Mol. Biol. 2012, 78, 223–246.
- Seki, M.; Narusaka, M.; Abe, H.; Kasuga, M.; Yamaguchi-Shinozaki, K.; Carninci, P.; Hayashizaki, Y.; Shinozaki, K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 2001, 13, 61–72.
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309.
- Zhu, J.; Verslues, P.E.; Zheng, X.; Lee, B.-H.; Zhan, X.; Manabe, Y.; Sokolchik, I.; Zhu, Y.; Dong, C.-H.; Zhu, J.-K. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc. Natl. Acad. Sci. USA 2005, 102, 9966–9971.
- Hihara, Y.; Kamei, A.; Kanehisa, M.; Kaplan, A.; Ikeuchi, M. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 2001, 13, 793–806.
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751.
- Hwang, E.-W.; Kim, K.-A.; Park, S.-C.; Jeong, M.-J.; Byun, M.-O.; Kwon, H.-B. Expression profiles of hot pepper (Capsicum annuum) genes under cold stress conditions. J. Biosci. 2005, 30, 657–667.
- da Silva, J.A.T.; Yücel, M. Revealing response of plants to biotic and abiotic stresses with microarray technology. Genes Genomes Genom. 2008, 2, 14–48.
- Ravikumar, G.; Manimaran, P.; Voleti, S.R.; Subrahmanyam, D.; Sundaram, R.M.; Bansal, K.C.; Viraktamath, B.C.; Balachandran, S.M. Stress-inducible expression of AtDREB1A transcription factor greatly improves drought stress tolerance in transgenic indica rice. Transgenic Res. 2014, 23, 421–439.
- Leviatan, N.; Alkan, N.; Leshkowitz, D.; Fluhr, R. Genome-wide survey of cold stress regulated alternative splicing in Arabidopsis thaliana with tiling microarray. PLoS ONE 2013, 8, e66511.
- Oono, Y.; Seki, M.; Nanjo, T.; Narusaka, M.; Fujita, M.; Satoh, R.; Satou, M.; Sakurai, T.; Ishida, J.; Akiyama, K. Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca. 7000 full-length cDNA microarray. Plant J. 2003, 34, 868–887.
- Maruyama, K.; Sakuma, Y.; Kasuga, M.; Ito, Y.; Seki, M.; Goda, H.; Shimada, Y.; Yoshida, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004, 38, 982–993.
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992.
- Song, X.; Li, Y.; Hou, X. Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 573.
- Lotfi, A.; Pervaiz, T.; Jiu, S.; Faghihi, F.; Jahanbakhshian, Z.; Khorzoghi, E.G.; Fang, J. Role of microRNAs and their target genes in salinity response in plants. Plant Growth Regul. 2017, 82, 377–390.
- Lata, R.; Chowdhury, S.; Gond, S.K.; White, J.F., Jr. Induction of abiotic stress tolerance in plants by endophytic microbes. Lett. Appl. Microbiol. 2018, 66, 268–276.
- Kumar, R.; Mustafiz, A.; Sahoo, K.K.; Sharma, V.; Samanta, S.; Sopory, S.K.; Pareek, A.; Singla-Pareek, S.L. Functional screening of cDNA library from a salt tolerant rice genotype Pokkali identifies mannose-1-phosphate guanyl transferase gene (OsMPG1) as a key member of salinity stress response. Plant Mol. Biol. 2012, 79, 555–568.
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467.
- Hu, Y.; Burucs, Z.; von Tucher, S.; Schmidhalter, U. Short-term effects of drought and salinity on mineral nutrient distribution along growing leaves of maize seedlings. Environ. Exp. Bot. 2007, 60, 268–275.
- Mangrauthia, S.K.; Agarwal, S.; Sailaja, B.; Madhav, M.S.; Voleti, S. MicroRNAs and their role in salt stress response in plants. In Salt Stress in Plants; Springer: Berlin/Heidelberg, Germany, 2013; pp. 15–46.
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132.
- Ouyang, B.; Yang, T.; Li, H.; Zhang, L.; Zhang, Y.; Zhang, J.; Fei, Z.; Ye, Z. Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. J. Exp. Bot. 2007, 58, 507–520.
- Sahi, C.; Singh, A.; Kumar, K.; Blumwald, E.; Grover, A. Salt stress response in rice: Genetics, molecular biology, and comparative genomics. Funct. Integr. Genom. 2006, 6, 263–284.
- Zhu, Y.N.; Shi, D.Q.; Ruan, M.B.; Zhang, L.L.; Meng, Z.H.; Liu, J.; Yang, W.C. Transcriptome analysis reveals crosstalk of responsive genes to multiple abiotic stresses in cotton (Gossypium hirsutum L.). PLoS ONE 2013, 8, e80218.
- Liu, H.-H.; Tian, X.; Li, Y.-J.; Wu, C.-A.; Zheng, C.-C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. Rna 2008, 14, 836–843.
- Chan, Z.; Bigelow, P.J.; Loescher, W.; Grumet, R. Comparison of salt stress resistance genes in transgenic Arabidopsis thaliana indicates that extent of transcriptomic change may not predict secondary phenotypic or fitness effects. Plant Biotechnol. J. 2012, 10, 284–300.
- Khraiwesh, B.; Zhu, J.-K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 137–148.
- Kumari, S.; Singh, P.; Singla-Pareek, S.L.; Pareek, A. Heterologous expression of a salinity and developmentally regulated rice cyclophilin gene (OsCyp2) in E. coli and S. cerevisiae confers tolerance towards multiple abiotic stresses. Mol. Biotechnol. 2009, 42, 195–204.
- Wang, C.; Han, J.; Liu, C.; Kibet, K.N.; Kayesh, E.; Shangguan, L.; Li, X.; Fang, J. Identification of microRNAs from Amur grape (Vitis amurensis Rupr.) by deep sequencing and analysis of microRNA variations with bioinformatics. BMC Genom. 2012, 13, 122.
- Wu, T.; Yang, C.; Ding, B.; Feng, Z.; Wang, Q.; He, J.; Tong, J.; Xiao, L.; Jiang, L.; Wan, J. Microarray-based gene expression analysis of strong seed dormancy in rice cv. N22 and less dormant mutant derivatives. Plant Physiol. Biochem. 2016, 99, 27–38.
- Kawasaki, S.; Borchert, C.; Deyholos, M.; Wang, H.; Brazille, S.; Kawai, K.; Galbraith, D.; Bohnert, H.J. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 2001, 13, 889–905.
- Kong, W.; Zhong, H.; Gong, Z.; Fang, X.; Sun, T.; Deng, X.; Li, Y. Meta-analysis of salt stress transcriptome responses in different rice genotypes at the seedling stage. Plants 2019, 8, 64.
- Trindade, I.; Capitão, C.; Dalmay, T.; Fevereiro, M.P.; Dos Santos, D.M. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2010, 231, 705–716.
- Wang, T.; Chen, L.; Zhao, M.; Tian, Q.; Zhang, W.-H. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genom. 2011, 12, 1–11.
- Mantri, N.; Basker, N.; Ford, R.; Pang, E.; Pardeshi, V. The role of micro-ribonucleic acids in legumes with a focus on abiotic stress response. Plant Genome 2013, 6.
- Li, H.; Dong, Y.; Yin, H.; Wang, N.; Yang, J.; Liu, X.; Wang, Y.; Wu, J.; Li, X. Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biol. 2011, 11, 1–12.
- Dong, Z.; Shi, L.; Wang, Y.; Chen, L.; Cai, Z.; Wang, Y.; Jin, J.; Li, X. Identification and dynamic regulation of microRNAs involved in salt stress responses in functional soybean nodules by high-throughput sequencing. Int. J. Mol. Sci. 2013, 14, 2717–2738.
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273.
- Chen, L.; Heikkinen, L.; Wang, C.; Yang, Y.; Sun, H.; Wong, G. Trends in the development of miRNA bioinformatics tools. Brief Bioinform. 2019, 20, 1836–1852.
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009, 19, 1175–1183.
- Bhattacharya, A.; Ziebarth, J.D.; Cui, Y. PolymiRTS Database 3.0: Linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014, 42, D86–D91.
- Cho, S.; Jang, I.; Jun, Y.; Yoon, S.; Ko, M.; Kwon, Y.; Choi, I.; Chang, H.; Ryu, D.; Lee, B.; et al. MiRGator v3.0: A microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013, 41, D257.
- Wang, X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics 2016, 32, 1316–1322.
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370.
- Preusse, M.; Theis, F.J.; Mueller, N.S. miTALOS v2: Analyzing Tissue Specific microRNA Function. PLoS ONE 2016, 11, e0151771.
- Loher, P.; Rigoutsos, I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics 2012, 28, 3322–3323.
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54.
- Zhou, K.R.; Liu, S.; Sun, W.J.; Zheng, L.L.; Zhou, H.; Yang, J.H.; Qu, L.H. ChIPBase v2.0: Decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017, 45, D43–D50.
- Fromm, B.; Billipp, T.; Peck, L.E.; Johansen, M.; Tarver, J.E.; King, B.L.; Newcomb, J.M.; Sempere, L.F.; Flatmark, K.; Hovig, E.; et al. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu. Rev. Genet. 2015, 49, 213–242.
- Ma, X.; Liu, C.; Gu, L.; Mo, B.; Cao, X.; Chen, X. TarHunter, a tool for predicting conserved microRNA targets and target mimics in plants. Bioinformatics 2018, 34, 1574–1576.
- Haunsberger, S.J.; Connolly, N.M.; Prehn, J.H. miRNAmeConverter: An R/bioconductor package for translating mature miRNA names to different miRBase versions. Bioinformatics 2017, 33, 592–593.
- Page, G.P.; Coulibaly, I. Bioinformatic tools for inferring functional information from plant microarray data: Tools for the first steps. Int. J. Plant Genom. 2008, 2008, 147563.
- Demeter, J.; Beauheim, C.; Gollub, J.; Hernandez-Boussard, T.; Jin, H.; Maier, D.; Matese, J.C.; Nitzberg, M.; Wymore, F.; Zachariah, Z.K.; et al. The Stanford Microarray Database: Implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007, 35, D766–D770.
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121.
- Alonso-Betanzos, A.; Bolón-Canedo, V.; Morán-Fernández, L.; Sánchez-Maroño, N. A Review of Microarray Datasets: Where to Find Them and Specific Characteristics. Methods Mol. Biol. 2019, 1986, 65–85.
- Mehta, J.P.; Rani, S. Software and tools for microarray data analysis. Methods Mol. Biol. 2011, 784, 41–53.
- Kawaura, K.; Mochida, K.; Ogihara, Y. Genome-wide analysis for identification of salt-responsive genes in common wheat. Funct. Integr. Genom. 2008, 8, 277–286.
- Fernandez, P.; Di Rienzo, J.; Fernandez, L.; Hopp, H.E.; Paniego, N.; Heinz, R.A. Transcriptomic identification of candidate genes involved in sunflower responses to chilling and salt stresses based on cDNA microarray analysis. BMC Plant Biol. 2008, 8, 11.
- Singla-Pareek, S.L.; Yadav, S.K.; Pareek, A.; Reddy, M.K.; Sopory, S.K. Enhancing salt tolerance in a crop plant by overexpression of glyoxalase II. Transgenic Res. 2008, 17, 171–180.
- Uddin, M.I.; Qi, Y.; Yamada, S.; Shibuya, I.; Deng, X.P.; Kwak, S.S.; Kaminaka, H.; Tanaka, K. Overexpression of a new rice vacuolar antiporter regulating protein OsARP improves salt tolerance in tobacco. Plant Cell Physiol. 2008, 49, 880–890.
- Sakamoto, K.; Göransson, O.; Hardie, D.G.; Alessi, D.R. Activity of LKB1 and AMPK-related kinases in skeletal muscle: Effects of contraction, phenformin, and AICAR. Am. J. Physiol. Endocrinol. Metab. 2004, 287, E310–E317.
- Hichri, I.; Muhovski, Y.; Žižková, E.; Dobrev, P.I.; Gharbi, E.; Franco-Zorrilla, J.M.; Lopez-Vidriero, I.; Solano, R.; Clippe, A.; Errachid, A. The Solanum lycopersicum WRKY3 transcription factor SlWRKY3 is involved in salt stress tolerance in tomato. Front. Plant Sci. 2017, 8, 1343.
- Mukhopadhyay, S.; Mandal, S.K.; Bhaduri, S.; Armstrong, W.H. Manganese clusters with relevance to photosystem II. Chem. Rev. 2004, 104, 3981–4026.
- De Lorenzo, L.; Merchan, F.; Blanchet, S.; Megías, M.; Frugier, F.; Crespi, M.; Sousa, C. Differential expression of the TFIIIA regulatory pathway in response to salt stress between Medicago truncatula genotypes. Plant Physiol. 2007, 145, 1521–1532.
- Liu, M.; Shi, J.; Lu, C. Identification of stress-responsive genes in Ammopiptanthus mongolicususing ESTs generated from cold-and drought-stressed seedlings. BMC Plant Biol. 2013, 13, 88.
- Havaux, M.; Guedeney, G.; Hagemann, M.; Yeremenko, N.; Matthijs, H.C.; Jeanjean, R. The chlorophyll-binding protein IsiA is inducible by high light and protects the cyanobacterium Synechocystis PCC6803 from photooxidative stress. FEBS Lett. 2005, 579, 2289–2293.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
2 times
(View History)
Update Date:
13 Jan 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




