Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Guosheng Xie | + 2235 word(s) | 2235 | 2021-10-22 08:27:54 | | | |
| 2 | Jessie Wu | Meta information modification | 2235 | 2021-11-03 04:30:23 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Xie, G. Cold Stress in Arabidopsis and Rice. Encyclopedia. Available online: https://encyclopedia.pub/entry/15619 (accessed on 07 February 2026).
Xie G. Cold Stress in Arabidopsis and Rice. Encyclopedia. Available at: https://encyclopedia.pub/entry/15619. Accessed February 07, 2026.
Xie, Guosheng. "Cold Stress in Arabidopsis and Rice" Encyclopedia, https://encyclopedia.pub/entry/15619 (accessed February 07, 2026).
Xie, G. (2021, November 02). Cold Stress in Arabidopsis and Rice. In Encyclopedia. https://encyclopedia.pub/entry/15619
Xie, Guosheng. "Cold Stress in Arabidopsis and Rice." Encyclopedia. Web. 02 November, 2021.
Copy Citation
Cold stress, including freezing stress and chilling stress, is one of the major environmental factors that limit the growth and productivity of plants. As a temperate dicot model plant species, Arabidopsis develops a capability to freezing tolerance through cold acclimation. The past decades have witnessed a deep understanding of mechanisms underlying cold stress signal perception, transduction, and freezing tolerance in Arabidopsis. In contrast, a monocot cereal model plant species derived from tropical and subtropical origins, rice, is very sensitive to chilling stress and has evolved a different mechanism for chilling stress signaling and response.
cold stress
signal perception
Arabidopsis
rice
1. Cold Stress Perceptions at Plasma Membrane (PM) in Arabidopsis and Rice
One of the major consequences of the temperature downshift is a decrease in membrane fluidity affecting membrane-associated cellular functions, and the PM is proposed as a primary sensor of low-temperature stress [1][2][3]. The feature of primary perception of temperature in plants has been proposed [4]. Different microdomains with lipid raft formation and composition, including sphingolipids in the PM, are responsible for sensing the particular temperature ranges [5][1]. Many putative calcium channels, PM-bound G-protein associated receptors, plasma membrane-localized receptor-like kinases (RLKs) have been identified as cold sensors in plants. Calcium channels responsible for Ca2+ influx have been considered a major sensor class for low temperature [6][7][8]. Through the membrane rigidification-activated mechano-sensitive or ligand-activated Ca2+ channels, cold stress induces a transient Ca2+ influx into the cytosol (Figure 1a). Two Arabidopsis calcium-permeable mechano-sensitive channels, AtMCA1 and AtMCA2, are involved in a cold-induced increase in [Ca2+]cyt and cold stress tolerance [9]. A cold sensor OsCOLD1 is the novel PM and endoplasmic reticulum (ER)-located protein, which interacted with α subunit 1 of the G protein (RGA1), enhancing the calcium transients in the cytosol in cold signal transduction in rice [10]. Two cyclic nucleotide-gated channels, OsCNGC14 and OsCNGC16, mediate the calcium signaling and promote chilling tolerance in rice seedlings. Their homologous proteins AtCNGC2 and AtCNGC4 in Arabidopsis promote chilling growth and freezing tolerance [11]. OsCNGC9 positively regulates chilling tolerance by mediating cytoplasmic calcium signaling in rice [12] (Figure 1b). Therefore, calcium channels play a central role in cold stress sensing in Arabidopsis and rice.
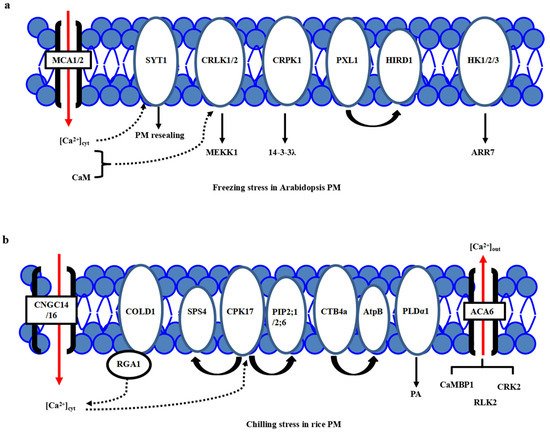
Figure 1. Plasma membrane-localized proteins perceive the cold stress signals in Arabidopsis and rice. (a) In Arabidopsis, freezing stress initiates the PM rigidification, PM-associated calcium channels MCA1/2, calcium sensor SYT1 and kinases including CRLK1, AtHK1/2/3, and CRPK1, as well as PM-localized PXL1, participate in primary cold stress sensing and perception. (b) In rice, chilling stress initiates the PM rigidification, many PM-associated proteins, including calcium channels ACA6, CNGC14/16, phospholipidase PLDα1, aquaporin proteins PIP2;1/PIP2;6, G-protein-associated cold sensor COLD1 and kinases GT4a and CPK17, participate in primary cold stress sensing and perception. However, a specific calcium channel for calcium influx is still not known.
On the other hand, a new calcium sensor synaptotagmin without the EF-hand motif, AtSYT1, localized to the PM and ER, participates in the exocytosis process in the calcium-dependent pathway under freezing stress in Arabidopsis [13] (Figure 1a). In rice, thirteen SYT homologous N-terminal-TM-C2 domain proteins (OsNTMC2) have been annotated [14]. However, the function of OsNTMC2 in vesicle trafficking and PM repair in cold stress response awaits further investigations.
Many receptor-like protein kinases, such as two-component histidine kinases, RLKs, and G-protein associated kinases, have played pivotal roles in cold stress sensing in Arabidopsis and rice. Two-component signaling systems, AHK2/3, AHP2/3/5, and ARR7, mediate the cold stress signaling through inhibiting ABA signaling [15][16]. Besides, AtCRLK1 binds to calcium and calmodulin (CaM), interacts with phosphorylates AtMEKK1 in freezing signaling and tolerance [17]. Moreover, AtCRPK1 phosphorylates 14-3-3λ which shuttles from the cytosol to the nucleus, then interacts with and destabilizes the CBFs in freezing stress tolerance [18]. In addition, AtPXL1 interacts with and phosphorylates histidine-rich dehydrin1 (AtHIRD1) and a light-harvesting protein complex I (AtLHCA1) to positively regulate cold and heat stress tolerances during the germination stage [19].
In rice, there are several identified PM-localized kinases involved in cold stress perception. OsACA6, a PM Ca2+-ATPase, interacts with CaM-binding protein OsCaMBP1, calcium-dependent protein kinase (CDPK)-related kinase OsCRK2, and receptor-like kinase (RLK) OsRLK2 [20]. PM-localized OsCPK17 interacts with and phosphorylates the sucrose-phosphate synthase OsSPS4 and aquaporin OsPIP2;1/OsPIP2;6, can enhance the cold stress tolerance in rice [21](Figure 1b). CTB4a, a conserved leucine-rich repeat receptor-like kinase, interacts with a beta subunit of adenosine triphosphate (ATP) synthase AtpB and improves the yield under cold stress [22]. Interestingly, the protein level of phospholipase Dα1 (OsPLDα1) increases at one minute after cold treatment. It activates OsMPK6 and OsSIZ1, followed by the regulations of OsDREB1s expression in cold signaling [23] (Figure 1b). Therefore, there is much convergence of primary PM-located protein kinases in cold stress perceptions between Arabidopsis and rice.
2. Cold Stress Signal Transduction Mechanisms in Arabidopsis and Rice
Following the cold stress perceptions, cold stress signal transduction events occur in the cytosol and nucleus of plant cells. The second messengers, such as Ca2+ and reactive oxygen species (ROS), transmit the external cold signals to intracellular signaling systems. Progress has been made in calcium signaling, phospholipid signaling, MAPK cascade signaling, and ROS signaling in the past decades. Here, we compare the recent advances in signal transduction pathways of freezing stress in Arabidopsis and chilling stress in rice (Figure 2 and Figure 3), highlighting the divergence and convergence in cold stress in both plant species.
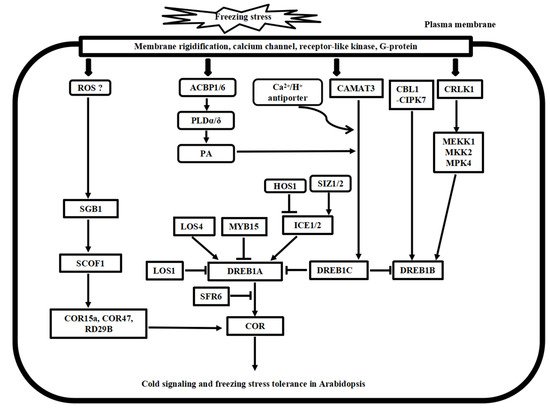
Figure 2. Putative model of cold stress signaling networks toward freezing stress tolerance in Arabidopsis. The cold-induced calcium signature in the cytosol is recognized by the calcium sensor proteins, including CaM, CDPK, CBL1/CIPK7, and CAMAT3, as well as the bZIP transcription factor SGB1 pathway. In addition, CRLK1-MEK1-MKK1/2-MPK4/6 cascade, ROS signaling, and phospholipid signaling work together to regulate cold stress signaling, and many ICE1-DREB transcription activators and repressors have been identified to regulate the COR gene expressions, finally leading to freezing tolerance in Arabidopsis.
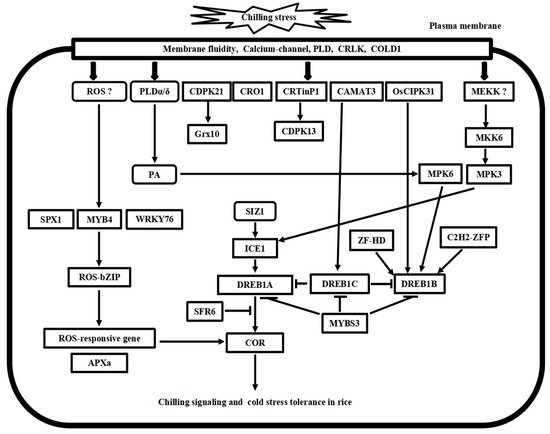
Figure 3. Putative model of chilling stress signaling networks toward cold stress tolerance in rice. There are at least four chilling stress signaling pathways in rice. MYB4-ROS-bZIP cascade is involved in the ROS signaling process. MKK6-MP3 cascade and phospholipid signaling work with calcium signaling concomitantly in chilling stress signaling in rice. ICE1-DREB transcriptional regulatory cascade is conserved in the Arabidopsis and rice. Furthermore, these pathways’ upstream and downstream signal transducer proteins play cooperative and regulatory roles in cold stress tolerance in rice.
2.1. Calcium Signaling
Calcium influx into the cytosol is an early event in cold stress [24][3]. This transient elevation in calcium concentration is also called intracellular calcium signature. Calcium influx is primarily sensed by the calcium sensor proteins, containing the helix-loop-helix domain with the EF-hand motif. In plants, calcium sensors include four major classes: CaM/CaM-like protein (CML), calcium-dependent protein kinase (CDPK or CPK), calcineurin B-like (CBL) protein, and CBL-interacting protein kinase (CIPK). In addition, a small annexin family has been identified as a calcium sensor to cold stress response in Arabidopsis.
In Arabidopsis, overexpression of AtCaM3 hinders the cold induction of RD29A and KIN1, and the AtCaM4 negatively regulates freezing tolerance by interacting with a CaM-binding protein PATL1 [25]. AtCBL1 interacts with AtCIPK7 and binds to the DREB core element of COR promoters to negatively regulate freezing tolerance [26]. CaM-binding transcription activator protein CAMTA3 binds to the conserved CG-1 element in the CBF2 promoter, regulating CBF2 expression in cold stress signaling [27]. A vacuolar Ca2+/H+ antiporter AtCAX1 enhances the DREB1 transcription in cold acclimation response [28]. Recently, an AtOST1-AtANN1cascade was found to regulate calcium signaling in the CBF1-dependent manner to enhance freezing tolerance in Arabidopsis [29]. This evidence demonstrated the negative and positive regulations of calcium sensors to freezing tolerance in Arabidopsis.
In rice, a CaM-like protein OsCML16 and its six putative targets have been identified to be involved in cold stress response in rice [30]. However, there is no report about the role of OsCaMs in cold stress signaling. OsCDPK7 enhances cold stress tolerance by the increased accumulation of a putative target gene rab16A [31][32]. OsCDPK13 enhances cold stress tolerance by activating a ubiquitin-like nuclear protein OsCRTintP1, calreticulin interacting protein 1 [33][34]. OsCPK24 interacts with and phosphorylates OsGrx10, a glutathione-dependent thioltransferase, in cold stress response [35]. Overexpression of OsCIPK3, a CBL-interacting protein kinase, improves cold stress tolerance [36]. It is worth mentioning that OsCIPK31 is strongly induced by cold and salt stress and interacts with AtCBL3, suggesting the convergence of CBL/CIPK pathways in cold stress signaling Arabidopsis and rice [37]. As described above, the CaM-associated signaling pathways in cold stress signaling wait for further confirmations in Arabidopsis and rice.
2.2. Phospholipid Signaling
An increasing number of studies have shown that the metabolism of the membrane lipids plays an important role in the temperature stress response in plants. In Arabidopsis, in a few seconds after cold exposure, diacylglycerol kinase (DGK) is activated to converse diacylglycerol (DAG) into phosphatidic acid (PA), followed by a change in membrane fluidity [38]. Overexpression of a PM-bound phospholipase gene PLDδ enhances freezing tolerance in rice seedlings [39]. Suppressed expression of PLDα1 results in a significant increase in freezing tolerance [40]. Acyl-coenzyme A: diacylglycerol acyltransferase DGAT1 enhances freezing tolerance via CBF2 regulon and NADPH oxidase RbohD (respiratory burst oxidase homolog D)-dependent H2O2 production in Arabidopsis [41]. The acyl-coenzyme A-binding protein (ACBP) family has six members (AtACBP1-6) in Arabidopsis. Overexpression of AtACBP6 enhances freezing tolerance by activating PLDδ to decrease phosphatidylcholine (PC) levels and accumulate PA [42]. Overexpression of AtACBP1 increases freezing sensitivity via the expression of PLDα1 and PLDδ and maintains a membrane-associated PA pool [43]. Further, a temperature-induced lipid pathway has been demonstrated. The FAD2, FAD5 and ACT1 have been identified as the key enzymes in influencing fatty acid flux between the eukaryotic and prokaryotic pathways cold stress response in Arabidopsis [44].
In plants, glycerol-3-phosphate acyltransferase (GPAT) of chloroplasts is a key enzyme to catalyze transferring the acyl group of acyl-(acyl-carrier-protein) (ACP) into the sn-1 position of glycerol 3-phosphate in the first step of glycerolipid biosynthesis in chloroplasts. Ectopic overexpressing of AtGPAT in rice largely induces the unsaturation of fatty acids and chilling tolerance of photosynthesis under low temperature [45]. In rice, the ω-3 fatty acid (FA) desaturase (FAD8) mutant does not acclimate to cold stress [46]. OsPLDα1 increases the levels of PA that bind to OsMPK6 in cold signaling and tolerance [23]. Interestingly, comparative glycerolipidomics analysis of freezing stress (−6 °C and −12 °C) in Arabidopsis and chilling stress (4 °C and 10 °C) in rice has illustrated that Arabidopsis has a higher double bond index (DBI) and lower average acyl chain length (ACL) than rice under cold stress condition [47]. Accordingly, glycerolipid metabolism and signaling show great potentials in applying cold stress tolerance engineering in Arabidopsis and rice.
2.3. MAPK Cascade Signaling
In plants, the MAPK cascade consists of three sequentially phosphorylating and activating components, a MAP kinase kinase kinase (MEKK/MAPKKK), a MAP kinase kinase (MEK/MAPKK), and a MAP kinase (MPK/MAPK). MAPKs phosphorylate various downstream substrates, including transcription factors, protein kinases, phospholipases, and cytoskeleton-associated proteins, finally leading to the activation of specific gene expressions under stress conditions [48].
In Arabidopsis, MAPKKK protein AtANP1 initiates a phosphorylation cascade with AtMPK3 under cold stress [49]. The complete cascade AtCRLK1-AtMEKK1-AtMKK2-AtMPK4/6 has been established to positively regulate freezing tolerance [50]. Recently, AtMPK6 is found to phosphorylate AtMYB15 to reduce the binding affinity of AtCBF3 and freezing tolerance [51]. AtMPK3 and AtMPK6 phosphorylate AtICE1 to promote its degradation, thereby negatively regulate freezing tolerance [52]. AtMPK6 phosphorylates AtMYB5 to positively regulate freezing tolerance [51]. However, the AtMEKK1-AtMKK1/2-AtMPK4 cascade promotes freezing tolerance by antagonizing the AtMPK3/6 pathway [53]. These results indicate that AtMPK3, AtMPK4, and AtMPK6 proteins cooperatively regulate freezing tolerance in Arabidopsis.
In rice, there is not identified complete MAPK pathway involved in cold stress signaling until now. Our previous study established that the OsMKK6-OsMPK3 cascade modulates chilling signaling and tolerance in rice [54]. OsMPK3 phosphorylates and stabilizes OsICE1, which directly transactivates the expression of OsTPP1, thereby positively regulating chilling tolerance [55]. Moreover, PA binds to OsMPK6 and mediates chilling stress signaling and tolerance [23]. Therefore, these results have demonstrated a divergence in MAPK signaling pathways and regulation network in cold stress response in Arabidopsis and rice.
2.4. ROS Signaling
Under cold stress, excess ROS is produced and brings about oxidative damage and cold stress response in plant cells. In Arabidopsis, AtMEKK1-AtMKK2-AtMPK4/AtMPK6 cascade regulates the ROS-scavenging enzymes to maintain redox homeostasis under cold stress [50]. Overexpression of a ROS-regulated C2H2 zinc finger transcription factor AtZAT12 decreases the expressions of AtCBF1/2/3 genes under cold stress [56]. In addition, AtHAP5A, a heme-associated protein, positively modulates the freezing resistance by binding AtXTH21 and inhibits ROS accumulation under freezing stress [57]. Stromal and thylakoid-bound ascorbate peroxidases sAPX and tAPX trigger COR15A, PAL1, and CHS expressions under cold stress [58]. AtTrx-h2 regulates the expressions of COR genes under freezing stress [59].
H2O2 levels are increased within 1.5 h of 10 °C stress in rice seedlings [60]. A subset of 121 early-response genes was upregulated during the initial 24 h of 10 °C stress [61]; Among them, four are transcription factor genes, including ROS-bZIP1 and asl/ocs-like element-containing genes. A hypothetical model of ROS-mediated regulon (ROS-bZIP-as1/ocs) is assembled independent of CBF/DREB- or ABA-mediated regulons in cold stress response [60][61]. Comparative metabolomics analysis of indica (9311) and japonica (Nipponbare) varieties revealed a ROS-dominated dynamic model involved in chilling stress adaptation and tolerance in rice [62]. Overexpression of OsZFP245 enhances cold stress tolerance by regulating proline levels and ROS-scavenging activities in rice seedlings [63]. Overexpressing OsAPX1 prevents the over-accumulation of H2O2 and reduces lipid peroxidation in the spikelet tissues at the booting stage of rice [64]. Natural variation reveals that OsSAP16 controls low-temperature germination in rice [65]. Therefore, there exist specific and different pathways of ROS-mediated cold signaling in rice.
References
- Murata, N.; Los, D.A. Membrane fluidity and temperature perception. Plant. Physiol. 1997, 115, 875–879.
- Sharma, A.; Isogai, M.; Yamamoto, T.; Sakaguchi, K.; Hashimoto, J.; Komatsu, S. A novel interaction between calreticulin and ubiquitin-like nuclear protein in rice. Plant. Cell Physiol. 2004, 45, 684–692.
- Penfield, S. Temperature perception and signal transduction in plants. New Phytol. 2008, 179, 615–628.
- McClung, C.R.; Davis, S.J. Ambient thermometers in plants: From physiological outputs towards mechanisms of thermal sensing. Curr. Biol. 2010, 20, R1086–R1092.
- Knight, M.R.; Knight, H. Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 2012, 195, 737–751.
- Monroy, A.F.; Dhindsa, R.S. Low-temperature signal transduction: Induction of cold acclimation-specific genes of alfalfa by calcium at 25 degrees c. Plant. Cell 1995, 7, 321–331.
- Knight, H.; Trewavas, A.J.; Knight, M.R. Cold calcium signaling in arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant. Cell 1996, 8, 489–503.
- Kiegle, E.; Moore, C.A.; Haseloff, J.; Tester, M.A.; Knight, M.R. Cell-type-specific calcium responses to drought, salt and cold in the arabidopsis root. Plant. J. 2000, 23, 267–278.
- Mori, K.; Renhu, N.; Naito, M.; Nakamura, A.; Shiba, H.; Yamamoto, T.; Suzaki, T.; Iida, H.; Miura, K. Ca(2+)-permeable mechanosensitive channels mca1 and mca2 mediate cold-induced cytosolic ca(2+) increase and cold tolerance in arabidopsis. Sci. Rep. 2018, 8, 550.
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D. Cold1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221.
- Cui, Y.; Lu, S.; Li, Z.; Cheng, J.; Hu, P.; Zhu, T.; Wang, X.; Jin, M.; Wang, X.; Li, L.; et al. Cyclic nucleotide-gated ion channels 14 and 16 promote tolerance to heat and chilling in rice. Plant. Physiol. 2020, 183, 1794–1808.
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y.; et al. Transcriptional activation and phosphorylation of oscngc9 confer enhanced chilling tolerance in rice. Mol. Plant. 2021, 14, 315–329.
- Yamazaki, T.; Kawamura, Y.; Minami, A.; Uemura, M. Calcium-dependent freezing tolerance in arabidopsis involves membrane resealing via synaptotagmin syt1. Plant. Cell 2008, 20, 3389–3404.
- Huang, R.; Zhao, J.; Liu, J.; Wang, Y.; Han, S.; Zhao, H. Genome-wide analysis and expression profiles of ntmc2 family genes in oryza sativa. Gene 2017, 637, 130–137.
- Jeon, J.; Kim, J. Arabidopsis response regulator1 and arabidopsis histidine phosphotransfer protein2 (ahp2), ahp3, and ahp5 function in cold signaling. Plant. Physiol. 2013, 161, 408–424.
- Jeon, J.; Kim, N.Y.; Kim, S.; Kang, N.Y.; Novak, O.; Ku, S.J.; Cho, C.; Lee, D.J.; Lee, E.J.; Strnad, M.; et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in arabidopsis. J. Biol. Chem. 2010, 285, 23371–23386.
- Yang, T.; Shad Ali, G.; Yang, L.; Du, L.; Reddy, A.S.; Poovaiah, B.W. Calcium/calmodulin-regulated receptor-like kinase crlk1 interacts with mekk1 in plants. Plant. Signal. Behav. 2010, 5, 991–994.
- Liu, Z.; Jia, Y.; Ding, Y.; Shi, Y.; Li, Z.; Guo, Y.; Gong, Z.; Yang, S. Plasma membrane crpk1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune cbf signaling during cold response. Mol. Cell 2017, 66, 117–128.e5.
- Jung, C.G.; Hwang, S.G.; Park, Y.C.; Park, H.M.; Kim, D.S.; Park, D.H.; Jang, C.S. Molecular characterization of the cold- and heat-induced arabidopsis pxl1 gene and its potential role in transduction pathways under temperature fluctuations. J. Plant. Physiol. 2015, 176, 138–146.
- Kamrul Huda, K.M.; Akhter Banu, M.S.; Yadav, S.; Sahoo, R.K.; Tuteja, R.; Tuteja, N. Salinity and drought tolerant osaca6 enhances cold tolerance in transgenic tobacco by interacting with stress-inducible proteins. Plant. Physiol. Biochem. 2014, 82, 229–238.
- Almadanim, M.C.; Alexandre, B.M.; Rosa, M.T.; Sapeta, H.; Leitão, A.E.; Ramalho, J.C.; Lam, T.T.; Negrão, A.; Abreu, I.A.; Oliveira, M.M. Rice calcium-dependent protein kinase oscpk17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant. Cell Environ. 2017, 40, 1197–1213.
- Zhang, Z.; Li, J.; Pan, Y.; Zhou, L.; Shi, H.; Zeng, Y.; Guo, H.; Yang, S.; Zheng, W.; Yu, J.; et al. Natural variation in ctb4a enhances rice adaptation to cold habitats. Nat. Commun. 2017, 8, 14788.
- Huo, C.; Zhang, B.; Wang, H.; Wang, F.; Liu, M.; Gao, Y.; Zhang, W.; Deng, Z.; Sun, D.; Tang, W. Comparative study of early cold-regulated proteins by two-dimensional difference gel electrophoresis reveals a key role for phospholipase dalpha1 in mediating cold acclimation signaling pathway in rice. Mol. Cell Proteom. 2016, 15, 1397–1411.
- Chinnusamy, V.; Zhu, J.K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 2010, 639, 39–55.
- Chu, M.; Li, J.; Zhang, J.; Shen, S.; Li, C.; Gao, Y.; Zhang, S. Atcam4 interacts with a sec14-like protein, patl1, to regulate freezing tolerance in arabidopsis in a cbf-independent manner. J. Exp. Bot. 2018, 69, 5241–5253.
- Huang, C.; Ding, S.; Zhang, H.; Du, H.; An, L. Cipk7 is involved in cold response by interacting with cbl1 in arabidopsis thaliana. Plant. Sci. 2011, 181, 57–64.
- Doherty, C.J.; Van Buskirk, H.A.; Myers, S.J.; Thomashow, M.F. Roles for arabidopsis camta transcription factors in cold-regulated gene expression and freezing tolerance. Plant. Cell 2009, 21, 972–984.
- Catala, R.; Santos, E.; Alonso, J.M.; Ecker, J.R.; Martinez-Zapater, J.M.; Salinas, J. Mutations in the ca2+/h+ transporter cax1 increase cbf/dreb1 expression and the cold-acclimation response in arabidopsis. Plant. Cell 2003, 15, 2940–2951.
- Liu, Q.; Ding, Y.; Shi, Y.; Ma, L.; Wang, Y.; Song, C.; Wilkins, K.A.; Davies, J.M.; Knight, H.; Knight, M.R.; et al. The calcium transporter annexin1 mediates cold-induced calcium signaling and freezing tolerance in plants. EMBO J. 2021, 40, e104559.
- Yang, J.; Liu, S.; Ji, L.; Tang, X.; Zhu, Y.; Xie, G. Identification of novel oscml16 target proteins and differential expression analysis under abiotic stresses in rice. J. Plant. Physiol. 2020, 249, 153165.
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Over-expression of a single ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant. J. 2000, 23, 319–327.
- Saijo, Y.; Kinoshita, N.; Ishiyama, K.; Hata, S.; Kyozuka, J.; Hayakawa, T.; Nakamura, T.; Shimamoto, K.; Yamaya, T.; Izui, K. A ca(2+)-dependent protein kinase that endows rice plants with cold- and salt-stress tolerance functions in vascular bundles. Plant. Cell Physiol. 2001, 42, 1228–1233.
- Abbasi, F.; Onodera, H.; Toki, S.; Tanaka, H.; Komatsu, S. Oscdpk13, a calcium-dependent protein kinase gene from rice, is induced by cold and gibberellin in rice leaf sheath. Plant. Mol. Biol. 2004, 55, 541–552.
- Komatsu, S.; Yang, G.; Khan, M.; Onodera, H.; Toki, S.; Yamaguchi, M. Over-expression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Mol. Genet. Genom. 2007, 277, 713–723.
- Liu, Y.; Xu, C.; Zhu, Y.; Zhang, L.; Chen, T.; Zhou, F.; Chen, H.; Lin, Y. The calcium-dependent kinase oscpk24 functions in cold stress responses in rice. J. Integr. Plant. Biol. 2018, 60, 173–188.
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of stress-responsive cipk genes in rice for stress tolerance improvement. Plant. Physiol. 2007, 144, 1416–1428.
- Kim, K.N.; Lee, J.S.; Han, H.; Choi, S.A.; Go, S.J.; Yoon, I.S. Isolation and characterization of a novel rice ca2+-regulated protein kinase gene involved in responses to diverse signals including cold, light, cytokinins, sugars and salts. Plant. Mol. Biol. 2003, 52, 1191–1202.
- Vaultier, M.N.; Cantrel, C.; Vergnolle, C.; Justin, A.M.; Demandre, C.; Benhassaine-Kesri, G.; Cicek, D.; Zachowski, A.; Ruelland, E. Desaturase mutants reveal that membrane rigidification acts as a cold perception mechanism upstream of the diacylglycerol kinase pathway in arabidopsis cells. FEBS Lett. 2006, 580, 4218–4223.
- Li, W.; Li, M.; Zhang, W.; Welti, R.; Wang, X. The plasma membrane-bound phospholipase ddelta enhances freezing tolerance in arabidopsis thaliana. Nat. Biotechnol. 2004, 22, 427–433.
- Rajashekar, C.B.; Zhou, H.E.; Zhang, Y.; Li, W.; Wang, X. Suppression of phospholipase dalpha1 induces freezing tolerance in arabidopsis: Response of cold-responsive genes and osmolyte accumulation. J. Plant. Physiol. 2006, 163, 916–926.
- Tan, W.J.; Yang, Y.C.; Zhou, Y.; Huang, L.P.; Xu, L.; Chen, Q.F.; Yu, L.J.; Xiao, S. Diacylglycerol acyltransferase and diacylglycerol kinase modulate triacylglycerol and phosphatidic acid production in the plant response to freezing stress. Plant. Physiol. 2018, 177, 1303–1318.
- Chen, Q.F.; Xiao, S.; Chye, M.L. Overexpression of the arabidopsis 10-kilodalton acyl-coenzyme a-binding protein acbp6 enhances freezing tolerance. Plant. Physiol. 2008, 148, 304–315.
- Du, Z.Y.; Chen, M.X.; Chen, Q.F.; Gu, J.D.; Chye, M.L. Expression of arabidopsis acyl-coa-binding proteins atacbp1 and atacbp4 confers pb(ii) accumulation in brassica juncea roots. Plant. Cell Env. 2015, 38, 101–117.
- Li, Q.; Zheng, Q.; Shen, W.; Cram, D.; Fowler, D.B.; Wei, Y.; Zou, J. Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. Plant. Cell 2015, 27, 86–103.
- Ariizumi, T.; Kishitani, S.; Inatsugi, R.; Nishida, I.; Murata, N.; Toriyama, K. An increase in unsaturation of fatty acids in phosphatidylglycerol from leaves improves the rates of photosynthesis and growth at low temperatures in transgenic rice seedlings. Plant. Cell Physiol. 2002, 43, 751–758.
- Tovuu, A.; Zulfugarov, I.S.; Wu, G.X.; Kang, I.S.; Kim, C.; Moon, B.Y.; An, G.; Lee, C.H. Rice mutants deficient in omega-3 fatty acid desaturase (fad8) fail to acclimate to cold temperatures. Plant. Physiol. Biochem. 2016, 109, 525–535.
- Zheng, G.; Li, L.; Li, W. Glycerolipidome responses to freezing- and chilling-induced injuries: Examples in arabidopsis and rice. BMC Plant. Biol. 2016, 16, 70.
- Hamel, L.P.; Nicole, M.C.; Sritubtim, S.; Morency, M.J.; Ellis, M.; Ehlting, J.; Beaudoin, N.; Barbazuk, B.; Klessig, D.; Lee, J.; et al. Ancient signals: Comparative genomics of plant mapk and mapkk gene families. Trends Plant. Sci. 2006, 11, 192–198.
- Kovtun, Y.; Chiu, W.L.; Tena, G.; Sheen, J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 2000, 97, 2940–2945.
- Teige, M.; Scheikl, E.; Eulgem, T.; Doczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The mkk2 pathway mediates cold and salt stress signaling in arabidopsis. Mol. Cell 2004, 15, 141–152.
- Kim, S.H.; Kim, H.S.; Bahk, S.; An, J.; Yoo, Y.; Kim, J.Y.; Chung, W.S. Phosphorylation of the transcriptional repressor myb15 by mitogen-activated protein kinase 6 is required for freezing tolerance in arabidopsis. Nucleic Acids Res. 2017, 45, 6613–6627.
- Zhao, C.; Wang, P.; Si, T.; Hsu, C.C.; Wang, L.; Zayed, O.; Yu, Z.; Zhu, Y.; Dong, J.; Tao, W.A.; et al. Map kinase cascades regulate the cold response by modulating ice1 protein stability. Dev. Cell 2017, 43, 618–629.e5.
- Li, H.; Ding, Y.; Shi, Y.; Zhang, X.; Zhang, S.; Gong, Z.; Yang, S. Mpk3- and mpk6-mediated ice1 phosphorylation negatively regulates ice1 stability and freezing tolerance in arabidopsis. Dev. Cell 2017, 43, 630–642.e4.
- Xie, G.; Kato, H.; Imai, R. Biochemical identification of the osmkk6-osmpk3 signalling pathway for chilling stress tolerance in rice. Biochem. J. 2012, 443, 95–102.
- Zhang, Z.; Li, J.; Li, F.; Liu, H.; Yang, W.; Chong, K.; Xu, Y. Osmapk3 phosphorylates osbhlh002/osice1 and inhibits its ubiquitination to activate ostpp1 and enhances rice chilling tolerance. Dev. Cell 2017, 43, 731–743.e5.
- Vogel, J.T.; Zarka, D.G.; Van Buskirk, H.A.; Fowler, S.G.; Thomashow, M.F. Roles of the cbf2 and zat12 transcription factors in configuring the low temperature transcriptome of arabidopsis. Plant. J. 2005, 41, 195–211.
- Shi, H.; Ye, T.; Zhong, B.; Liu, X.; Jin, R.; Chan, Z. Athap5a modulates freezing stress resistance in arabidopsis through binding to ccaat motif of atxth21. New Phytol. 2014, 203, 554–567.
- van Buer, J.; Cvetkovic, J.; Baier, M. Cold regulation of plastid ascorbate peroxidases serves as a priming hub controlling ros signaling in arabidopsis thaliana. BMC Plant. Biol. 2016, 16, 163.
- Park, J.H.; Lee, E.S.; Chae, H.B.; Paeng, S.K.; Wi, S.D.; Bae, S.B.; Thi Phan, K.A.; Lee, S.Y. Disulfide reductase activity of thioredoxin-h2 imparts cold tolerance in arabidopsis. Biochem. Biophys. Res. Commun. 2021, 568, 124–130.
- Yun, K.Y.; Park, M.R.; Mohanty, B.; Herath, V.; Xu, F.; Mauleon, R.; Wijaya, E.; Bajic, V.B.; Bruskiewich, R.; de Los Reyes, B.G. Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant. Biol. 2010, 10, 16.
- Cheng, C.; Yun, K.Y.; Ressom, H.W.; Mohanty, B.; Bajic, V.B.; Jia, Y.; Yun, S.J.; de los Reyes, B.G. An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genom. 2007, 8, 175.
- Zhang, J.; Luo, W.; Zhao, Y.; Xu, Y.; Song, S.; Chong, K. Comparative metabolomic analysis reveals a reactive oxygen species-dominated dynamic model underlying chilling environment adaptation and tolerance in rice. New Phytol. 2016, 211, 1295–1310.
- Huang, J.; Sun, S.-J.; Xu, D.-Q.; Yang, X.; Bao, Y.-M.; Wang, Z.-F.; Tang, H.-J.; Zhang, H. Increased tolerance of rice to cold, drought and oxidative stresses mediated by the overexpression of a gene that encodes the zinc finger protein zfp245. Biochem. Biophys. Res. Commun. 2009, 389, 556–561.
- Sato, Y.; Masuta, Y.; Saito, K.; Murayama, S.; Ozawa, K. Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene, osapxa. Plant. Cell Rep. 2011, 30, 399–406.
- Wang, X.; Zou, B.; Shao, Q.; Cui, Y.; Lu, S.; Zhang, Y.; Huang, Q.; Huang, J.; Hua, J. Natural variation reveals that ossap16 controls low-temperature germination in rice. J. Exp. Bot. 2018, 69, 413–421.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.7K
Entry Collection:
Environmental Sciences
Revisions:
2 times
(View History)
Update Date:
03 Nov 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




