
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Fiona Errington-Mais | + 804 word(s) | 804 | 2020-11-05 09:14:58 | | | |
| 2 | Dean Liu | + 433 word(s) | 1237 | 2020-11-09 12:09:56 | | |
Video Upload Options
Oncolytic virotherapy (OVT) has received significant attention in recent years, especially since the approval of talimogene Laherparepvec (T-VEC) in 2015 by the Food and Drug administration (FDA). Mechanistic studies of oncolytic viruses (OVs) have revealed that most, if not all, OVs induce direct oncolysis and stimulate innate and adaptive anti-tumour immunity. With the advancement of tumour modelling, allowing characterisation of the effects of tumour microenvironment (TME) components and identification of the cellular mechanisms required for cell death (both direct oncolysis and anti-tumour immune responses), it is clear that a “one size fits all” approach is not applicable to all OVs, or indeed the same OV across different tumour types and disease locations. This article will provide an unbiased review of oncolytic reovirus (clinically formulated as pelareorep), including the molecular and cellular requirements for reovirus oncolysis and anti-tumour immunity, reports of pre-clinical efficacy and its overall clinical trajectory. Moreover, as it is now abundantly clear that the true potential of all OVs, including reovirus, will only be reached upon the development of synergistic combination strategies, reovirus combination therapeutics will be discussed, including the limitations and challenges that remain to harness the full potential of this promising therapeutic agent.
1. Introduction
Advancements in virology and molecular biology techniques over recent decades have allowed us to exploit the anti-tumour potential of oncolytic viruses (OVs) [1]. The unique ability of OVs to exploit oncogenic signalling pathways provides a significant advantage over traditional treatment modalities. OVs are specifically defined as viruses which: (i) preferentially infect and kill malignant cells through viral replication and oncolysis, and (ii) engage the immune system to promote anti-tumour immunity. Additional mechanisms of action have also been reported, including disruption of tumour-associated vasculature or stroma and modulation of the tumour microenvironment (TME)[2][3][4].
An array of OVs—naturally occurring, attenuated, and genetically modified—have been investigated in pre-clinical models and clinical trials but only two have received approval for clinical use: (i) a genetically engineered adenovirus H101, approved in China in 2005 [5], and (ii) the Food and Drug Administration (FDA)-approved talimogene laherparepvec (T-VEC)—a herpes simplex virus type 1 (HSV-1) genetically engineered to limit neurovirulence and promote an immunostimulatory environment[6][7].
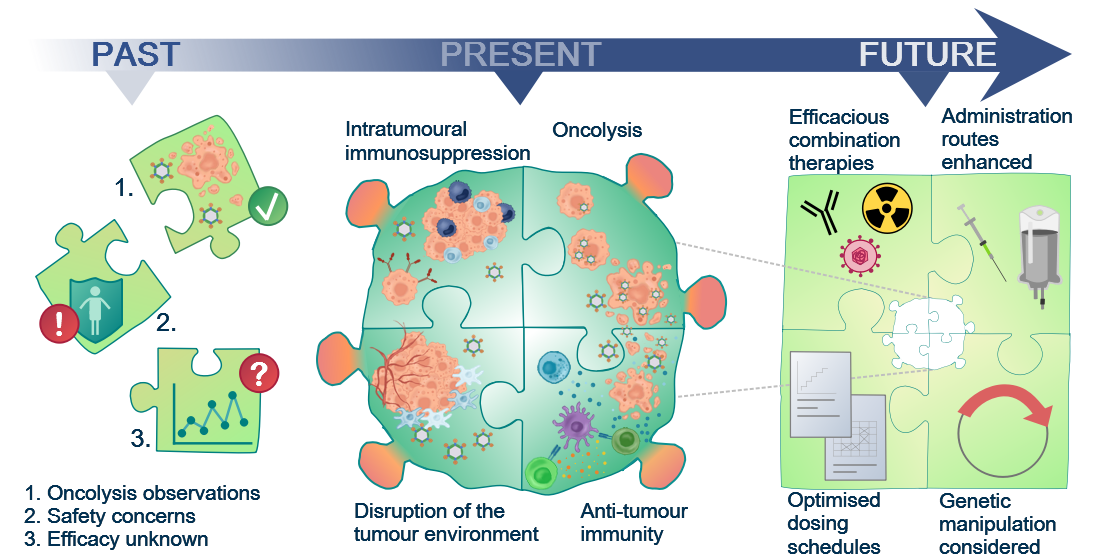
Figure 1. Although the oncolytic capacity of various viruses was noted already in the 1950s, development of oncolytic viruses (OVs) as therapeutic agents was limited by safety and efficacy concerns for a long time. With a greater understanding of virology and molecular techniques, the interest in OVs has peaked in recent years and a number of viruses, including reovirus, are now in clinical development. Today, we know that OVs exert their anti-tumour effects in several ways, e.g. direct oncolytic killing through replication in malignantly transformed cells; innate and adaptive anti-tumour immunity; disruption of the local tumour microenvironment sustaining the tumour; and intratumoural immunosuppressive effects, e.g. recruitment of regulatory T cells and enhanced expression of immune checkpoints on tumour cells. This knowledge will allow future initiatives to the identify the most efficacious combination therapies, optimised dosing schedules, and administration routes required for the success of reovirus as a therapeutic agent.
2. The Emergence of Reovirus as a Therapeutic Agent
The Reoviridae family of viruses has found hosts in mammals, fish, birds and plants [8][9]. Three serotypes of mammalian orthoreovirus have been identified: type one Lang, type two Jones, and type three Abney and Dearing [10]. Each differs in its in vivo tropism, despite a high degree of genetic similarity [11]. Type-specific diversity occurs in the S1 gene, encoding the outer capsid σ1 attachment protein, which has undergone significant evolutionary divergence[12]. Orthoreovirus type two Jones was the first serotype observed to replicate specifically in malignant cell lines[13]; however, it is the mammalian orthoreovirus type three Dearing strain (T3D)—now manufactured as pelareorep but previously known as Reolysin®—that has made progress as a therapeutic agent. Mammalian orthoreovirus T3D (hereafter referred to as reovirus) is typically isolated from human gastrointestinal and upper respiratory tracts[14][15]. In most individuals, infection proceeds asymptomatically causing mild enteric or respiratory illness in young children and being relatively non-pathogenic in adults, in line with its designation as a respiratory enteric orphan virus (reovirus)[10]. There have been sporadic reports of severe pathology associated with reovirus infection in infants and immunocompromised individuals [9][16][17][18][19][20][21] and more recently, reovirus has been implicated in coeliac disease by promoting a TH1 immune response, a response that bodes well for its use as an immunotherapeutic tool although oral delivery should be avoided to limit these potential unwanted side effects[22].
Reovirus is a non-enveloped, double-stranded (ds) RNA virus approximately 85 nm in diameter, with two concentric icosahedral protein capsids[23]. The outer and inner capsids protect the dsRNA genome which comprises 23.5 kbp in ten segments termed large (L1-3), medium (M1-3), or small (S1-4) according to size [23][24][25]. The gene segments encode eight structural proteins (λ1-3, µ1-2, and σ1-3) and the non-structural proteins, µNS and σNS[26]. μ1 and σ3 form part of the outer capsid, λ3 forms a subunit of the RNA polymerase and σ1 and λ2 are important for viral attachment, although σ1 initiates target cell entry[23]. The proteins also protect the virus from immune-surveillance by preventing a host anti-viral interferon (IFN) response; σ3 binds to dsRNA and prevents its binding to dsRNA-dependent protein kinase R (PKR; a dsRNA sensor)[27] and μNS sequesters the IFN transcription factor (interferon regulatory factor 3; IRF3) and inhibits its translocation to the nucleus[28].
3. Tumour Specificity and Replication
The reovirus life-cycle is shown in Figure 2. Viral entry occurs over multiple steps, the first being a low-affinity “tethering” of the reovirus σ1 protein to cell surface sialic acid[29][30]. Subsequently, σ1 engages junctional adhesion molecule A (JAM-A), the canonical reovirus receptor [31][32][33], which is ubiquitously expressed throughout the body and has several important roles in normal cellular processes including tight junction formation, leukocyte migration, and angiogenesis [34]. Fortuitously, JAM-A is also overexpressed in several cancers, including both haematological and solid malignancies[35][36][37][38][39][40][41]. Following reovirus engagement with JAM-A and receptor-mediated endocytosis, the viral particle undergoes acid-dependent cathepsin-mediated proteolysis within the endosome[42][43]to form an intermediate subviral particle (ISVP) characterised by the loss of σ3 and cleavage of µ1 [44]. The proteolytic uncoating, principally by cathepsins L and B, is critical for penetration of the endosome membrane by µ1; ISVPs undergo a conformational change causing autocleavage of µ1 into µ1N which triggers pore formation in the endocytic membrane[45] and delivers transcriptionally active reovirus into the cytosol[46][47] for replication. Capped, positive-sense single stranded (ss) RNA serves as mRNA for protein translation and provides a template for replication of nascent dsRNA genomes [48]. Transcription and translation occur in cytoplasmic “viral factories”[49][50], with packaging of the segmented genome into virions occurring concomitantly with RNA synthesis[51][52]. Viral egress can be non-cytolytic in the absence of transformation; however, the release of progeny virus is typically lytic in permissive, transformed cells[53][54].
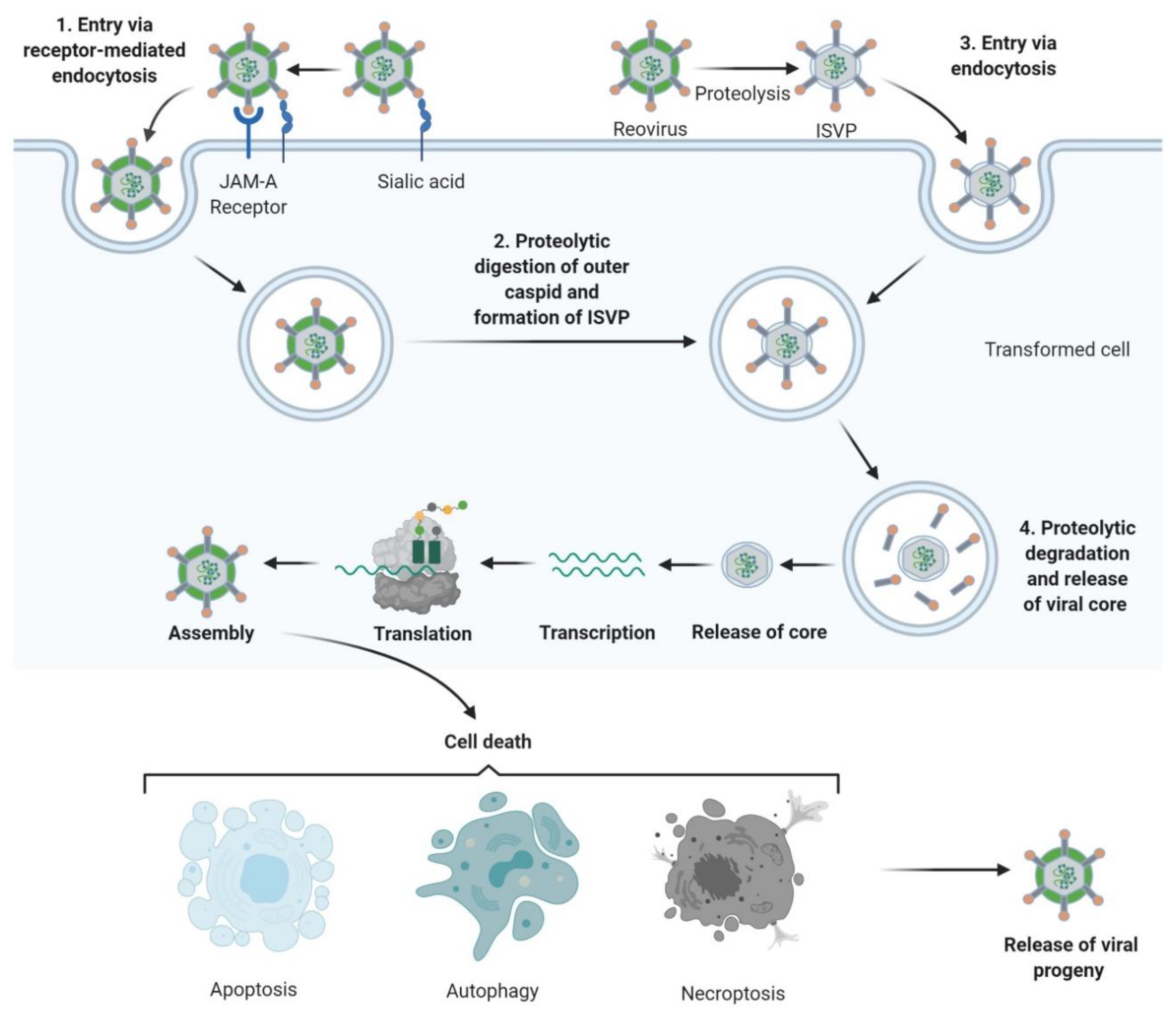
Figure 2. Reovirus replication: 1. Reovirus is first tethered via a weak interaction between σ1 and cell surface sialic acid; σ1 then binds with high affinity to junctional adhesion molecule A (JAM-A) resulting in internalization of the virus via receptor-mediated endocytosis. 2. Once internalized, the virus is transported to early and late endosomes where it undergoes proteolytic digestion to remove the outer capsid protein σ3 resulting in the formation of infectious subvirion particles (ISVPs). 3. Alternatively, ISVPs may be formed by extracellular proteases within the tumour environment allowing direct entry into cells via membrane penetration. 4. After further proteolytic degradation a transcriptionally active viral core is released into the cytoplasm. Transcription and translation occur ultimately leading to the assembly of new viral progeny, host cell death and progeny release. Figure created using Biorender (https://biorender.com/).
References
- Kelly, E.; Russell, S.J. History of oncolytic viruses: Genesis to genetic engineering. Mol. Ther. J. Am. Soc. Gene Ther. 2007, 15, 651–659.
- Breitbach, C.J.; Arulanandam, R.; De Silva, N.; Thorne, S.H.; Patt, R.; Daneshmand, M.; Moon, A.; Ilkow, C.; Burke, J.; Hwang, T.H.; et al. Oncolytic vaccinia virus disrupts tumor-associated vasculature in humans. Cancer Res. 2013, 73, 1265–1275.
- Ilkow, C.S.; Marguerie, M.; Batenchuk, C.; Mayer, J.; Ben Neriah, D.; Cousineau, S.; Falls, T.; Jennings, V.A.; Boileau, M.; Bellamy, D.; et al. Reciprocal cellular cross-talk within the tumor microenvironment promotes oncolytic virus activity. Nat. Med. 2015, 21, 530–536.
- Bauzon, M.; Hermiston, T. Armed therapeutic viruses—A disruptive therapy on the horizon of cancer immunotherapy. Front. Immunol. 2014, 5, 74.
- Garber, K. China approves world’s first oncolytic virus therapy for cancer treatment. J. Natl. Cancer Inst. 2006, 98, 298–300.
- Randazzo, B.P.; Tal-Singer, R.; Zabolotny, J.M.; Kesari, S.; Fraser, N.W. Herpes simplex virus 1716, an ICP 34.5 null mutant, is unable to replicate in CV-1 cells due to a translational block that can be overcome by coinfection with SV40. J. Gen. Virol 1997, 78 Pt 12, 3333–3339.
- Liu, B.L.; Robinson, M.; Han, Z.Q.; Branston, R.H.; English, C.; Reay, P.; McGrath, Y.; Thomas, S.K.; Thornton, M.; Bullock, P.; et al. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 2003, 10, 292–303.
- Lelli, D.; Beato, M.S.; Cavicchio, L.; Lavazza, A.; Chiapponi, C.; Leopardi, S.; Baioni, L.; De Benedictis, P.; Moreno, A. First identification of mammalian orthoreovirus type 3 in diarrheic pigs in Europe. Virol. J. 2016, 13, 139.
- Steyer, A.; Gutierrez-Aguire, I.; Kolenc, M.; Koren, S.; Kutnjak, D.; Pokorn, M.; Poljsak-Prijatelj, M.; Racki, N.; Ravnikar, M.; Sagadin, M.; et al. High similarity of novel orthoreovirus detected in a child hospitalized with acute gastroenteritis to mammalian orthoreoviruses found in bats in Europe. J. Clin. Microbiol. 2013, 51, 3818–3825.
- Rosen, L.; Hovis, J.F.; Mastrota, F.M.; Bell, J.A.; Huebner, R.J. Observations on a newly recognized virus (Abney) of the reovirus family. Am. J. Hyg. 1960, 71, 258–265.
- Gaillard, R.K., Jr.; Joklik, W.K. Quantitation of the relatedness of reovirus serotypes 1, 2, and 3 at the gene level. Virology 1982, 123, 152–164.
- Cashdollar, L.W.; Chmelo, R.A.; Wiener, J.R.; Joklik, W.K. Sequences of the S1 genes of the three serotypes of reovirus. Proc. Natl. Acad. Sci. USA 1985, 82, 24–28.
- Hashiro, G.; Loh, P.C.; Yau, J.T. The preferential cytotoxicity of reovirus for certain transformed cell lines. Arch. Virol. 1977, 54, 307–315.
- Ramos-Alvarez, M.; Sabin, A.B. Characteristics of poliomyelitis and other enteric viruses recovered in tissue culture from healthy American children. Proc. Soc. Exp. Biol. Med. 1954, 87, 655–661.
- Ramos-Alvarez, M.; Sabin, A.B. Enteropathogenic viruses and bacteria; role in summer diarrheal diseases of infancy and early childhood. J. Am. Med. Assoc. 1958, 167, 147–156.
- Chua, K.B.; Crameri, G.; Hyatt, A.; Yu, M.; Tompang, M.R.; Rosli, J.; McEachern, J.; Crameri, S.; Kumarasamy, V.; Eaton, B.T.; et al. A previously unknown reovirus of bat origin is associated with an acute respiratory disease in humans. Proc. Natl. Acad. Sci. USA 2007, 104, 11424–11429.
- Tillotson, J.R.; Lerner, A.M. Reovirus type 3 associated with fatal pneumonia. N. Engl. J. Med. 1967, 276, 1060–1063.
- Ouattara, L.A.; Barin, F.; Barthez, M.A.; Bonnaud, B.; Roingeard, P.; Goudeau, A.; Castelnau, P.; Vernet, G.; Paranhos-Baccala, G.; Komurian-Pradel, F. Novel human reovirus isolated from children with acute necrotizing encephalopathy. Emerg. Infect. Dis. 2011, 17, 1436–1444.
- Tyler, K.L.; Barton, E.S.; Ibach, M.L.; Robinson, C.; Campbell, J.A.; O’Donnell, S.M.; Valyi-Nagy, T.; Clarke, P.; Wetzel, J.D.; Dermody, T.S. Isolation and molecular characterization of a novel type 3 reovirus from a child with meningitis. J. Infect. Dis. 2004, 189, 1664–1675.
- Morecki, R.; Glaser, J.H.; Cho, S.; Balistreri, W.F.; Horwitz, M.S. Biliary atresia and reovirus type 3 infection. N. Engl. J. Med. 1982, 307, 481–484.
- Richardson, S.C.; Bishop, R.F.; Smith, A.L. Reovirus serotype 3 infection in infants with extrahepatic biliary atresia or neonatal hepatitis. J. Gastroenterol. Hepatol. 1994, 9, 264–268.
- Bouziat, R.; Hinterleitner, R.; Brown, J.J.; Stencel-Baerenwald, J.E.; Ikizler, M.; Mayassi, T.; Meisel, M.; Kim, S.M.; Discepolo, V.; Pruijssers, A.J.; et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 2017, 356, 44–50.
- Dermody, T.; Parker, J.; Sherry, B. Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott, Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2, pp. 1304–1346.
- Lemay, G. Transcriptional and translational events during reovirus infection. Biochem. Cell Biol. 1988, 66, 803–812.
- Shatkin, A.J.; Sipe, J.D.; Loh, P. Separation of ten reovirus genome segments by polyacrylamide gel electrophoresis. J. Virol. 1968, 2, 986–991.
- Chandran, K.; Zhang, X.; Olson, N.H.; Walker, S.B.; Chappell, J.D.; Dermody, T.S.; Baker, T.S.; Nibert, M.L. Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein. J. Virol. 2001, 75, 5335–5342.
- Yue, Z.; Shatkin, A.J. Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology 1997, 234, 364–371.
- Stanifer, M.L.; Kischnick, C.; Rippert, A.; Albrecht, D.; Boulant, S. Reovirus inhibits interferon production by sequestering IRF3 into viral factories. Sci. Rep. 2017, 7, 10873.
- Barton, E.S.; Connolly, J.L.; Forrest, J.C.; Chappell, J.D.; Dermody, T.S. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem. 2001, 276, 2200–2211.
- Chappell, J.D.; Duong, J.L.; Wright, B.W.; Dermody, T.S. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol. 2000, 74, 8472–8479.
- Antar, A.A.; Konopka, J.L.; Campbell, J.A.; Henry, R.A.; Perdigoto, A.L.; Carter, B.D.; Pozzi, A.; Abel, T.W.; Dermody, T.S. Junctional adhesion molecule-A is required for hematogenous dissemination of reovirus. Cell Host Microbe 2009, 5, 59–71.
- Barton, E.S.; Forrest, J.C.; Connolly, J.L.; Chappell, J.D.; Liu, Y.; Schnell, F.J.; Nusrat, A.; Parkos, C.A.; Dermody, T.S. Junction adhesion molecule is a receptor for reovirus. Cell 2001, 104, 441–451.
- Shmulevitz, M.; Gujar, S.A.; Ahn, D.G.; Mohamed, A.; Lee, P.W. Reovirus variants with mutations in genome segments S1 and L2 exhibit enhanced virion infectivity and superior oncolysis. J. Virol. 2012, 86, 7403–7413.
- Sugano, Y.; Takeuchi, M.; Hirata, A.; Matsushita, H.; Kitamura, T.; Tanaka, M.; Miyajima, A. Junctional adhesion molecule-A, JAM-A, is a novel cell-surface marker for long-term repopulating hematopoietic stem cells. Blood 2008, 111, 1167–1172.
- Parrish, C.; Scott, G.B.; Migneco, G.; Scott, K.J.; Steele, L.P.; Ilett, E.; West, E.J.; Hall, K.; Selby, P.J.; Buchanan, D.; et al. Oncolytic reovirus enhances rituximab-mediated antibody-dependent cellular cytotoxicity against chronic lymphocytic leukaemia. Leukemia 2015, 29, 1799–1810.
- Kelly, K.R.; Espitia, C.M.; Zhao, W.; Wendlandt, E.; Tricot, G.; Zhan, F.; Carew, J.S.; Nawrocki, S.T. Junctional adhesion molecule-A is overexpressed in advanced multiple myeloma and determines response to oncolytic reovirus. Oncotarget 2015, 6, 41275–41289.
- Hall, K.; Scott, K.J.; Rose, A.; Desborough, M.; Harrington, K.; Pandha, H.; Parrish, C.; Vile, R.; Coffey, M.; Bowen, D.; et al. Reovirus-mediated cytotoxicity and enhancement of innate immune responses against acute myeloid leukemia. BioRes. Open Access 2012, 1, 3–15.
- Xu, P.P.; Sun, Y.F.; Fang, Y.; Song, Q.; Yan, Z.X.; Chen, Y.; Jiang, X.F.; Fei, X.C.; Zhao, Y.; Leboeuf, C.; et al. JAM-A overexpression is related to disease progression in diffuse large B-cell lymphoma and downregulated by lenalidomide. Sci. Rep. 2017, 7, 7433.
- Zhao, C.; Lu, F.; Chen, H.; Zhao, X.; Sun, J.; Chen, H. Dysregulation of JAM-A plays an important role in human tumor progression. Int. J. Clin. Exp. Pathol. 2014, 7, 7242–7248.
- Zhang, M.; Luo, W.; Huang, B.; Liu, Z.; Sun, L.; Zhang, Q.; Qiu, X.; Xu, K.; Wang, E. Overexpression of JAM-A in non-small cell lung cancer correlates with tumor progression. PLoS ONE 2013, 8, e79173.
- McSherry, E.A.; McGee, S.F.; Jirstrom, K.; Doyle, E.M.; Brennan, D.J.; Landberg, G.; Dervan, P.A.; Hopkins, A.M.; Gallagher, W.M. JAM-A expression positively correlates with poor prognosis in breast cancer patients. Int. J. Cancer 2009, 125, 1343–1351.
- Alain, T.; Kim, T.S.; Lun, X.; Liacini, A.; Schiff, L.A.; Senger, D.L.; Forsyth, P.A. Proteolytic disassembly is a critical determinant for reovirus oncolysis. Mol. Ther. J. Am. Soc. Gene Ther. 2007, 15, 1512–1521.
- Golden, J.W.; Linke, J.; Schmechel, S.; Thoemke, K.; Schiff, L.A. Addition of exogenous protease facilitates reovirus infection in many restrictive cells. J. Virol. 2002, 76, 7430–7443.
- Ebert, D.H.; Deussing, J.; Peters, C.; Dermody, T.S. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 2002, 277, 24609–24617.
- Chandran, K.; Farsetta, D.L.; Nibert, M.L. Strategy for nonenveloped virus entry: A hydrophobic conformer of the reovirus membrane penetration protein micro 1 mediates membrane disruption. J. Virol. 2002, 76, 9920–9933.
- Nibert, M.L.; Odegard, A.L.; Agosto, M.A.; Chandran, K.; Schiff, L.A. Putative autocleavage of reovirus mu1 protein in concert with outer-capsid disassembly and activation for membrane permeabilization. J. Mol. Biol. 2005, 345, 461–474.
- Odegard, A.L.; Chandran, K.; Zhang, X.; Parker, J.S.; Baker, T.S.; Nibert, M.L. Putative autocleavage of outer capsid protein micro1, allowing release of myristoylated peptide micro1N during particle uncoating, is critical for cell entry by reovirus. J. Virol. 2004, 78, 8732–8745.
- Li, J.K.; Scheible, P.P.; Keene, J.D.; Joklik, W.K. The plus strand of reovirus gene S2 is identical with its in vitro transcript. Virology 1980, 105, 282–286.
- Fields, B.N.; Raine, C.S.; Baum, S.G. Temperature-sensitive mutants of reovirus type 3: Defects in viral maturation as studied by immunofluorescence and electron microscopy. Virology 1971, 43, 569–578.
- Kobayashi, T.; Chappell, J.D.; Danthi, P.; Dermody, T.S. Gene-specific inhibition of reovirus replication by RNA interference. J. Virol. 2006, 80, 9053–9063.
- Antczak, J.B.; Joklik, W.K. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology 1992, 187, 760–776.
- McDonald, S.M.; Patton, J.T. Assortment and packaging of the segmented rotavirus genome. Trends Microbiol. 2011, 19, 136–144.
- Connolly, J.L.; Barton, E.S.; Dermody, T.S. Reovirus binding to cell surface sialic acid potentiates virus-induced apoptosis. J. Virol. 2001, 75, 4029–4039.
- Lai, C.M.; Mainou, B.A.; Kim, K.S.; Dermody, T.S. Directional release of reovirus from the apical surface of polarized endothelial cells. mBio 2013, 4, e00049-13.




