
Video Upload Options
Asthma is a highly heterogeneous disease, but the pathogenesis of asthma is still unclear. It is well known that the airway inflammatory immune response is the pathological basis of asthma. Metabolomics is a systems biology method to analyze the difference of low molecular weight metabolites (<1.5 kDa) and explore the relationship between metabolic small molecules and pathophysiological changes of the organisms.
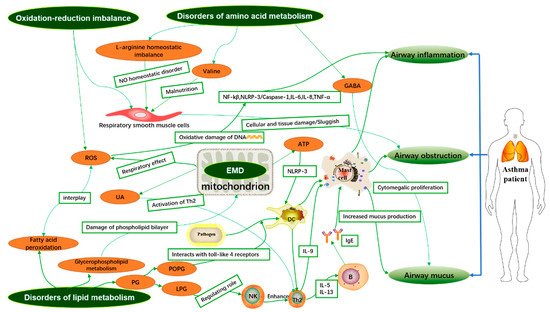
1. Introduction
2. Integrative Analysis of Asthma-Related Metabolites and Metabolic Pathways in Different Samples
| Author and Year | Subjects | Sample/Methods | Significant Metabolites | |
|---|---|---|---|---|
| Up | Down | |||
| Pang, Z. et al. (2018) [6] | eosinophilic asthmatics (EA, n = 13), noneosinophilic asthmatics (NEA, n = 16), and healthy controls (HC, n = 15) | Serum/UPLC-MS/MS | Monosaccharides, LysoPC(18:1), Retinyl ester, PC(18:1/2:0), LysoPC(o-18:0), Arachidonic acid, PE(18:3/14:0), PC(16:0/18:1) | Glycerophosphocholine, PS(18:0/22:5), Cholesterol glucuronide, Phytosphingosine, Sphinganine, LysoPC(p-18:1), Retinols, PC(20:4/16:1) |
| Guo, C. et al. (2021) [7] | 51 asthma patients and 9 healthy individuals | Serum/LC-MS | No report | SM 34:2, SM 38:1, SM 40:1 |
| Chiu, C.-Y. et al. (2020) [8] | Asthma (n = 28) and healthy controls (n = 26) | Plasma and urine/NMR | Histidine | 1-methylnicotinamide, trimethylamine N-oxide (TMAO) |
| Turi, K.N. et al. (2021) [9] | 600 infants from 3 independent cohorts | Plasma/LC-MS | Succinate, N-(2-furoyl)glycine | Iminodiacetate (IDA) |
| Jiang, T. et al. (2021) [10] | 28 healthy controls and 33 outpatients with asthma | Plasma/LC-MS/MS | Phosphatidylethanolamine (PE) (18:1p/22:6), PE (20:0/18:1), PE (38:1), sphingomyelin (SM) (d18:1/18:1), triglyceride (TG) (16:0/16:0/18:1) | Phosphatidylinositol (PI) (16:0/20:4), TG (17:0/18:1/18:1), phosphatidylglycerol (PG) (44:0), ceramide (Cer) (d16:0/27:2), lysophosphatidylcholine (LPC) (22:4) |
| Bian, X. et al. (2017) [11] | 15 healthy human and 15 asthma patients | Serum/UHPLC-Q-TOF-MS | Ursodeoxycholic acid, Deoxycholic acid, Isodeoxycholic acid, EPA | Palmitic acid, Lauric acid |
| Matysiak, J. et al. (2020) [12] | asthmatic children (n = 13) and the control group (n = 17) | Blood/LC-MS/MS | l-Arginine, Β-Alanine, Ƴ-Amino-N-Butyric Acid, l-Histidine, Hydroxy-l-Proline | d,l-Β-Aminoisobutyric Acid, Taurine, l-Tryptophan, l-Valine |
| Ghosh, N. et al. (2020) [13] | (i) controls = 33 (ii) asthma = 34 (iii) COPD = 30 and (iv) ACO = 35 | Serum/GC-MS | 2-palmitoylglycerol, cholesterol, serine, threonine, Ethanolamine, Glucose, Stearic acid, Linoleic acid, d-Mannose, Succinic acid | Lactic acid, 2-palmitoylglycerol |
| Liang, Y. et al. (2019) [14] | A total of 17 patients with mildly persistent asthma, 17 patients with stable COPD, and 15 healthy subjects | Serum/LC-MS | Hypoxanthine, P-chlorophenylalanine, Inosine, Theophylline, Bilirubin, Palmitic acid | l-Glutamine, Glycerophosphocholine, Succinate, Xanthine, Arachidonic Acid, l-Pyroglutamic acid, Indoxyl sulfate, l-Valine, l-Norleucine, l-Leucine, l-Phenylalanine |
| Chiu, C.-Y. et al. (2018) [15] | Asthma (n = 30) and healthy controls (n = 30) | Urine/NMR | Guanidoacetic acid | 1-methylnicotinamide, allantoin |
| Li, S. et al. (2020) [16] | Asthmatic children (n = 30) and healthy controls (n = 30) | Urine/GC-MS | l-allothreonine 1, stearic acid, succinic acid, 2-hydroxybutanoic acid, azelaic acid, gentiobiose 2, tyramine, leucine, d-altrose 1, d-erythrosphingosine 1, citraconic acid 4 | Valine, uric acid, methionine 1, 3,4-dihydroxycinnamic acid, purine riboside, malonic acid 1, cysteine, erythrose 1, lactamide 1 |
| Chawes, B.L. et al.(2018) [17] | 171 and 161 healthy neonates born from mothers with asthma | Urine/UPLC-MS | bile acid taurochenodeoxycholate-3-sulfate, fatty acid 3-hydroxytetradecanedioic acid | glucoronidated steroid compound |
| Carraro, S. et al. (2018) [18] | Children for transient wheezing (n = 16) and early-onset asthma (n = 16) | Urine/UPLC-MS | 4-(4-deoxy-α-d-gluc-4-enuronosyl)-d-galacturonate, Glutaric acid, 4-hydroxynonenal, Phosphatidyl glycerol, 3-methyluridine, Steroid O-sulfate, 5-hydroxy-l-tryptophan, 3-indoleacetic acid, Tiglylglycine, Indole, Cytosine, N-acetylputrescine, Indole-3-acetamide, 6-methyladenine, 5-methylcytosine, N-acryloylglycine, Hydroxyphenyllactic acid | Oxoadipic acid, (-)-epinephrine, l-tyrosine, 3-hydroxyhippuric acid, Benzoic acid,3-hydroxy-sebacic acid, Dihydroferulic acid 4-sulfate, p-cresol, Indolelactic acid, N-acetyl-l-phenylalanine, N2-acetyl-ornithine |
| Tao, J.-L. et al. (2019) [19] | Children for healthy control (n = 29), uncontrolled asthma (n = 37) or controlled asthma (n = 43) | Urine/GC-MS | Aspartic acid, Xanthosine, Hypoxanthine, N-acetylgalactosamine |
Stearic acid, Heptadecanoic acid, Uric acid, d-threitol |
| Adamko, D.J. et al. (2015) [20] | Adults with asthma (n = 58) and COPD (n = 24) | Urine/NMR | Glutamine, succinate, uracil, pantothenate | Arginine, dimethylamine, 3-Hydroxyisovalerate, betaine, choline, glucose, 1-methylnicotinamide |
| Ravi, A. et al. (2021) [21] | Healthy controls (n = 7) and patients with severe asthma (n = 9) | BECs/UPLC-MS | Phosphatidylcholines, lysophosphatidylcholines, lysophosphatidylethanolamines, bis(monoacylglycero)phosphates | No report |
| Chang-Chien, J. et al.(2020) [22] | stable asthma (n = 92) and non-asthmatic controls (n = 73) | EBC/NMR | lactate, formate, butyric acid, isobutyrate | No report |
| Ferraro, V.A. et al. (2020) [23] | asthmatic children (n = 26) and healthy children (n = 16) | EBC/UPLC-MS | 9-amino-nonanoic acid, 12-amino-dodecanoic acid, lactone of PGF-MUM, N-linoleoyl taurine, 17-phenoxy trinor PGF2α ethyl amide, lysoPC (18:2(9Z,12Z)) | No report |
| Kang, Y.P. et al. (2014) [24] | 38 asthma patients and 13 healthy subjects | BALF/HPLC-QTOF-MS | lysophosphatidylcholine (LPC), phosphatidylcholine (PC), phosphatidylglycerol (PG), phosphatidylserine (PS), sphingomyelin (SM), triglyceride (TG) | No report |
| Tian, M. et al. (2017) [25] | 15 healthy controls and 20 asthma patients | Sputum/UHPLC-QTOF-MS | Glycerol 1-stearate_1, 1-Hexadecanoyl-sn-glycerol_1, Cytidine 2′,3′-cyclic phosphate, 1-Hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-(1′-rac-glycerol), 1-Octadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoserine | His-Pro, Thr-Phe_1, Arg-Phe_1, Adenine_1, Phe-Tyr_1, Phe-Gln_1, Tyr-Ala_2, Phe-Ser_1, Urocanic acid |
Up: metabolites are higher in asthmatics than in normal people; Down: metabolites are lower in asthmatics than in normal people. UPLC-MS/MS: ultra-performance liquid chromatography-tandem mass spectrometry; LC-MS: liquid chromatography-mass spectrometry; NMR: nuclear magnetic resonance; LC-MS/MS: liquid chromatography-tandem mass spectrometry; GC-MS: Gas chromatography-mass spectrometry; UPLC-MS: ultra-performance liquid chromatography-mass spectrometry; HPLC-QTOF-MS: high performance liquid chromatography tandem quadrupole time-of-flight mass spectrometry; UHPLC-QTOF-MS: ultra-performance liquid chromatography tandem quadrupole time-of-flight mass spectrometry.
2.1. Analysis of Biomarkers Associated with Asthma in Blood/Serum/Plasma Samples
2.1.1. Phenotypic Identification and Treatment of Asthma
2.1.2. Diagnosis of Asthma
References
- Kaur, R.; Chupp, G. Phenotypes and endotypes of adult asthma: Moving toward precision medicine. J. Allergy Clin. Immunol. 2019, 144, 1–12.
- Szefler, S.J.; Wenzel, S.; Brown, R.; Erzurum, S.C.; Fahy, J.V.; Hamilton, R.G.; Hunt, J.F.; Kita, H.; Liu, A.H.; Panettieri, R.A.; et al. Asthma outcomes: Biomarkers. J. Allergy Clin. Immunol. 2012, 129, S9–S23.
- Aaron, S.D.; Boulet, L.P.; Reddel, H.; Gershon, A.S. Underdiagnosis and Overdiagnosis of Asthma. Am. J. Respir. Crit. Care Med. 2018, 198, 1012–1020.
- Kelly, R.S.; Dahlin, A.; McGeachie, M.J.; Qiu, W.; Sordillo, J.; Wan, E.S.; Wu, A.C.; Lasky-Su, J. Asthma Metabolomics and the Potential for Integrative Omics in Research and the Clinic. Chest 2017, 151, 262–277.
- Kuruvilla, M.E.; Lee, F.E.-H.; Lee, G.B. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin. Rev. Allergy Immunol. 2019, 56, 219–233.
- Pang, Z.; Wang, G.; Wang, C.; Zhang, W.; Liu, J.; Wang, F. Serum Metabolomics Analysis of Asthma in Different Inflammatory Phenotypes: A Cross-Sectional Study in Northeast China. BioMed Res. Int. 2018, 2018, 1–14.
- Guo, C.; Sun, L.; Zhang, L.; Dong, F.; Zhang, X.; Yao, L.; Chang, C. Serum sphingolipid profile in asthma. J. Leukoc. Biol. 2021, 110, 53–59.
- Chiu, C.-Y.; Cheng, M.-L.; Chiang, M.-H.; Wang, C.-J.; Tsai, M.-H.; Lin, G. Metabolomic Analysis Reveals Distinct Profiles in the Plasma and Urine Associated with IgE Reactions in Childhood Asthma. J. Clin. Med. 2020, 9, 887.
- Turi, K.N.; McKennan, C.; Gebretsadik, T.; Snyder, B.; Seroogy, C.M.; Lemanske, R.F.; Zoratti, E.; Havstad, S.; Ober, C.; Lynch, S.; et al. Unconjugated bilirubin is associated with protection from early-life wheeze and childhood asthma. J. Allergy Clin. Immunol. 2021.
- Jiang, T.; Dai, L.; Li, P.; Zhao, J.; Wang, X.; An, L.; Liu, M.; Wu, S.; Wang, Y.; Peng, Y.; et al. Lipid metabolism and identification of biomarkers in asthma by lipidomic analysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158853.
- Bian, X.; Sun, B.; Zheng, P.; Li, N.; Wu, J.-L. Derivatization enhanced separation and sensitivity of long chain-free fatty acids: Application to asthma using targeted and non-targeted liquid chromatography-mass spectrometry approach. Anal. Chim. Acta 2017, 989, 59–70.
- Matysiak, J.; Klupczynska, A.; Packi, K.; Mackowiak-Jakubowska, A.; Bręborowicz, A.; Pawlicka, O.; Olejniczak, K.; Kokot, Z.J.; Matysiak, J. Alterations in Serum-Free Amino Acid Profiles in Childhood Asthma. Int. J. Environ. Res. Public Health 2020, 17, 4758.
- Ghosh, N.; Choudhury, P.; Kaushik, S.R.; Arya, R.; Nanda, R.; Bhattacharyya, P.; Roychowdhury, S.; Banerjee, R.; Chaudhury, K. Metabolomic fingerprinting and systemic inflammatory profiling of asthma COPD overlap (ACO). Respir. Res. 2020, 21, 1–16.
- Liang, Y.; Gai, X.Y.; Chang, C.; Zhang, X.; Wang, J.; Li, T.T. Metabolomic Profiling Differences among Asthma, COPD, and Healthy Subjects: A LC-MS-based Metabolomic Analysis. Biomed. Environ. Sci. 2019, 32, 659–672.
- Chiu, C.-Y.; Lin, G.; Cheng, M.-L.; Chiang, M.-H.; Tsai, M.-H.; Su, K.-W.; Hua, M.-C.; Liao, S.-L.; Lai, S.-H.; Yao, T.-C.; et al. Longitudinal urinary metabolomic profiling reveals metabolites for asthma development in early childhood. Pediatr. Allergy Immunol. 2018, 29, 496–503.
- Li, S.; Liu, J.; Zhou, J.; Wang, Y.; Jin, F.; Chen, X.; Yang, J.; Chen, Z. Urinary Metabolomic Profiling Reveals Biological Pathways and Predictive Signatures Associated with Childhood Asthma. J. Asthma Allergy 2020, ume 13, 713–724.
- Chawes, B.L.; Giordano, G.; Pirillo, P.; Rago, D.; Rasmussen, M.A.; Stokholm, J.; Bønnelykke, K.; Bisgaard, H.; Baraldi, E. Neonatal Urine Metabolic Profiling and Development of Childhood Asthma. Metabolites 2019, 9, 185.
- Carraro, S.; Bozzetto, S.; Giordano, G.; El Mazloum, D.; Stocchero, M.; Pirillo, P.; Zanconato, S.; Baraldi, E. Wheezing preschool children with early-onset asthma reveal a specific metabolomic profile. Pediatr. Allergy Immunol. 2018, 29, 375–382.
- Tao, J.-L.; Chen, Y.-Z.; Dai, Q.-G.; Tian, M.; Wang, S.-C.; Shan, J.-J.; Ji, J.-J.; Lin, L.-L.; Li, W.-W.; Yuan, B. Urine metabolic profiles in paediatric asthma. Respirology 2019, 24, 572–581.
- Adamko, D.J.; Nair, P.; Mayers, I.; Tsuyuki, R.T.; Regush, S.; Rowe, B.H. Metabolomic profiling of asthma and chronic obstructive pulmonary disease: A pilot study differentiating diseases. J. Allergy Clin. Immunol. 2015, 136, 571–580.
- Ravi, A.; Goorsenberg, A.W.; Dijkhuis, A.; Dierdorp, B.S.; Dekker, T.; van Weeghel, M.; Piñeros, Y.S.S.; Shah, P.L.; Hacken, N.H.T.; Annema, J.T.; et al. Metabolic differences between bronchial epithelium from healthy individuals and patients with asthma and the effect of bronchial thermoplasty. J. Allergy Clin. Immunol. 2021, S0091–S6749, 00170-6.
- Chang-Chien, J.; Huang, H.; Tsai, H.; Lo, C.; Lin, W.; Tseng, Y.; Wang, S.; Ho, H.; Cheng, M.; Yao, T. Metabolomic differences of exhaled breath condensate among children with and without asthma. Pediatr. Allergy Immunol. 2021, 32, 264–272.
- Ferraro, V.A.; Carraro, S.; Pirillo, P.; Gucciardi, A.; Poloniato, G.; Stocchero, M.; Giordano, G.; Zanconato, S.; Baraldi, E. Breathomics in Asthmatic Children Treated with Inhaled Corticosteroids. Metabolites 2020, 10, 390.
- Kang, Y.P.; Lee, W.J.; Hong, J.Y.; Lee, S.B.; Park, J.H.; Kim, D.; Park, S.; Park, C.-S.; Park, S.-W.; Kwon, S.W. Novel Approach for Analysis of Bronchoalveolar Lavage Fluid (BALF) Using HPLC-QTOF-MS-Based Lipidomics: Lipid Levels in Asthmatics and Corticosteroid-Treated Asthmatic Patients. J. Proteome Res. 2014, 13, 3919–3929.
- Tian, M.; Chen, M.; Bao, Y.-L.; Xu, C.-D.; Qin, Q.-Z.; Zhang, W.-X.; He, Y.-T.; Shao, Q. Sputum metabolomic profiling of bronchial asthma based on quadruple time-of-flight mass spectrometry. Int. J. Clin. Exp. Pathol. 2017, 10, 10363–10373.
- Chung, K.F. Asthma phenotyping: A necessity for improved therapeutic precision and new targeted therapies. J. Intern. Med. 2016, 279, 192–204.
- Simpson, J.L.; Scott, R.; Boyle, M.J.; Gibson, P.G. Inflammatory subtypes in asthma: Assessment and identification using induced sputum. Respirology 2006, 11, 54–61.
- Mims, J.W. Asthma: Definitions and pathophysiology. Int. Forum Allergy Rhinol. 2015, 5, S2–S6.
- Fahy, J.V. Type 2 inflammation in asthma—Present in most, absent in many. Nat. Rev. Immunol. 2015, 15, 57–65.
- Bara, I.; Ozier, A.; De Lara, J.-M.T.; Marthan, R.; Berger, P. Pathophysiology of bronchial smooth muscle remodelling in asthma. Eur. Respir. J. 2010, 36, 1174–1184.
- Pite, H.; Morais-Almeida, M.; Rocha, S. Metabolomics in asthma. Curr. Opin. Pulm. Med. 2018, 24, 94–103.
- Gai, X.Y.; Zhang, L.J.; Chang, C.; Guo, C.L.; Abulikemu, M.; Li, W.X.; Wang, J.; Yao, W.Z.; Zhang, X. Metabolomic Anal-ysis of Serum Glycerophospholipid Levels in Eosinophilic and Neutrophilic Asthma. Biomed. Environ. Sci. 2019, 32, 96–106.
- Reinke, S.N.; Gallart-Ayala, H.; Gómez, C.; Checa, A.; Fauland, A.; Naz, S.; Kamleh, M.A.; Djukanović, R.; Hinks, T.S.; Wheelock, C.E. Metabolomics analysis identifies different metabotypes of asthma severity. Eur. Respir. J. 2017, 49, 1601740.
- Crestani, E.; Harb, H.; Charbonnier, L.-M.; Leirer, J.; Motsinger-Reif, A.; Rachid, R.; Phipatanakul, W.; Kaddurah-Daouk, R.; Chatila, T.A. Untargeted metabolomic profiling identifies disease-specific signatures in food allergy and asthma. J. Allergy Clin. Immunol. 2020, 145, 897–906.
- Chiu, C.; Chou, H.; Chang, L.; Fan, W.; Dinh, M.C.V.; Kuo, Y.; Chung, W.; Lai, H.; Hsieh, W.; Su, S. Integration of metagenomics-metabolomics reveals specific signatures and functions of airway microbiota in mite-sensitized childhood asthma. Allergy 2020, 75, 2846–2857.
- di Palmo, E.; Cantarelli, E.; Catelli, A.; Ricci, G.; Gallucci, M.; Miniaci, A.; Pession, A. The Predictive Role of Biomarkers and Genetics in Childhood Asthma Exacerbations. Int. J. Mol. Sci. 2021, 22, 4651.
- Rastogi, D.; Fraser, S.; Oh, J.; Huber, A.M.; Schulman, Y.; Bhagtani, R.H.; Khan, Z.S.; Tesfa, L.; Hall, C.; Macian, F. Inflammation, Metabolic Dysregulation, and Pulmonary Function among Obese Urban Adolescents with Asthma. Am. J. Respir. Crit. Care Med. 2015, 191, 149–160.
- Periyalil, H.A.; Gibson, P.G.; Wood, L.G. Immunometabolism in Obese Asthmatics: Are We There Yet? Nutrients 2013, 5, 3506–3530.
- Shore, S.A.; Cho, Y. Obesity and Asthma: Microbiome–Metabolome Interactions. Am. J. Respir. Cell Mol. Biol. 2016, 54, 609–617.
- Miethe, S.; Guarino, M.; Alhamdan, F.; Simon, H.-U.; Renz, H.; Dufour, J.-F.; Potaczek, D.P.; Garn, H. The effects of obesity on asthma: Immunometabolic links. Pol. Arch. Intern. Med. 2018, 128, 469–477.
- Liu, Y.; Zheng, J.; Zhang, H.P.; Zhang, X.; Wang, L.; Wood, L.; Wang, G. Obesity-Associated Metabolic Signatures Correlate to Clinical and Inflammatory Profiles of Asthma: A Pilot Study. Allergy Asthma Immunol. Res. 2018, 10, 628–647.
- Gomez-Llorente, M.A.; Martínez-Cañavate, A.; Chueca, N.; Rico, M.D.L.C.; Romero, R.; Anguita-Ruiz, A.; Aguilera, C.M.; Gil-Campos, M.; Mesa, M.D.; Khakimov, B.; et al. A Multi-Omics Approach Reveals New Signatures in Obese Allergic Asthmatic Children. Biomedicines 2020, 8, 359.
- Winnica, D.; Corey, C.; Mullett, S.; Reynolds, M.; Hill, G.; Wendell, S.; Que, L.; Holguin, F.; Shiva, S. Bioenergetic Differences in the Airway Epithelium of Lean Versus Obese Asthmatics Are Driven by Nitric Oxide and Reflected in Circulating Platelets. Antioxid. Redox Signal. 2019, 31, 673–686.
- Patel, M.; Pilcher, J.; Reddel, H.K.; Pritchard, A.; Corin, A.; Helm, C.; Tofield, C.; Shaw, D.; Black, P.; Weatherall, M.; et al. Metrics of salbutamol use as predictors of future adverse outcomes in asthma. Clin. Exp. Allergy 2013, 43, 1144–1151.
- McGeachie, M.J.; Dahlin, A.; Qiu, W.; Croteau-Chonka, D.C.; Savage, J.; Wu, A.C.; Wan, E.S.; Sordillo, J.E.; Al-Garawi, A.; Martinez, F.D.; et al. The metabolomics of asthma control: A promising link between genetics and disease. Immun. Inflamm. Dis. 2015, 3, 224–238.
- Yang, Y.; Uhlig, S. The role of sphingolipids in respiratory disease. Ther. Adv. Respir. Dis. 2011, 5, 325–344.
- Nixon, G.F. Sphingolipids in inflammation: Pathological implications and potential therapeutic targets. Br. J. Pharmacol. 2009, 158, 982–993.
- Yu, M.; Jia, H.-M.; Cui, F.-X.; Yang, Y.; Zhao, Y.; Yang, M.-H.; Zou, Z.-M. The Effect of Chinese Herbal Medicine Formula mKG on Allergic Asthma by Regulating Lung and Plasma Metabolic Alternations. Int. J. Mol. Sci. 2017, 18, 602.
- Ran, S.; Sun, F.; Song, Y.; Wang, X.; Hong, Y.; Han, Y. The Study of Dried Ginger and Linggan Wuwei Jiangxin Decoction Treatment of Cold Asthma Rats Using GC–MS Based Metabolomics. Front. Pharmacol. 2019, 10, 284.
- You, Y.-N.; Xing, Q.-Q.; Zhao, X.; Ji, J.-J.; Yan, H.; Zhou, T.; Dong, Y.-M.; Ren, L.-S.; Hou, S.-T.; Ding, Y.-Y. Gu-Ben-Fang-Xiao decoction modulates lipid metabolism by activating the AMPK pathway in asthma remission. Biomed. Pharmacother. 2021, 138, 111403.
- Bhavsar, P.; Hew, M.; Khorasani, N.; Torrego, A.; Barnes, P.J.; Adcock, I.; Chung, K.F. Relative corticosteroid insensitivity of alveolar macrophages in severe asthma compared with non-severe asthma. Thorax 2008, 63, 784–790.
- Pang, Z.; Ran, N.; Yuan, Y.; Wang, C.; Wang, G.; Lin, H.; Hsu, A.C.-Y.; Liu, J.; Wang, F. Phenotype-Specific Therapeutic Effect of Rhodiola wallichiana var. cholaensis Combined with Dexamethasone on Experimental Murine Asthma and Its Comprehensive Pharmacological Mechanism. Int. J. Mol. Sci. 2019, 20, 4216.
- Su, L.; Shi, L.; Liu, J.; Huang, L.; Huang, Y.; Nie, X. Metabolic profiling of asthma in mice and the interventional effects of SPA using liquid chromatography and Q-TOF mass spectrometry. Mol. BioSyst. 2017, 13, 1172–1181.
- Mochimaru, T.; Fukunaga, K.; Miyata, J.; Matsusaka, M.; Masaki, K.; Kabata, H.; Ueda, S.; Suzuki, Y.; Goto, T.; Urabe, D.; et al. 12-OH-17,18-Epoxyeicosatetraenoic acid alleviates eosinophilic airway inflammation in murine lungs. Allergy 2018, 73, 369–378.
- Comhair, S.A.A.; McDunn, J.; Bennett, C.; Fettig, J.; Erzurum, S.C.; Kalhan, S.C.; Fetig, J. Metabolomic Endotype of Asthma. J. Immunol. 2015, 195, 643–650.
- Izawa, K.; Isobe, M.; Matsukawa, T.; Ito, S.; Maehara, A.; Takahashi, M.; Yamanishi, Y.; Kaitani, A.; Oki, T.; Okumura, K.; et al. Sphingomyelin and ceramide are physiological ligands for human LMIR3/CD300f, inhibiting FcεRI-mediated mast cell activation. J. Allergy Clin. Immunol. 2014, 133, 270–273.e7.
- Schjødt, M.S.; Gürdeniz, G.; Chawes, B. The Metabolomics of Childhood Atopic Diseases: A Comprehensive Pathway-Specific Review. Metabolites 2020, 10, 511.
- Kelly, R.S.; Chawes, B.; Blighe, K.; Virkud, Y.V.; Croteau-Chonka, D.C.; McGeachie, M.J.; Clish, C.; Bullock, K.; Celedón, J.C.; Weiss, S.T.; et al. An Integrative Transcriptomic and Metabolomic Study of Lung Function in Children with Asthma. Chest 2018, 154, 335–348.
- Kelly, R.S.; Virkud, Y.; Giorgio, R.; Celedón, J.C.; Weiss, S.T.; Lasky-Su, J. Metabolomic profiling of lung function in Costa-Rican children with asthma. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1590–1595.
- Wang, S.; Tang, K.; Lu, Y.; Tian, Z.; Huang, Z.; Wang, M.; Zhao, J.; Xie, J. Revealing the role of glycerophospholipid metabolism in asthma through plasma lipidomics. Clin. Chim. Acta 2021, 513, 34–42.
- Funk, C.D. Prostaglandins and Leukotrienes: Advances in Eicosanoid Biology. Science 2001, 294, 1871–1875.
- Anthonisen, N.R.; Lindgren, P.G.; Tashkin, D.P.; Kanner, R.E.; Scanlon, P.D.; Connett, J.E. Bronchodilator response in the lung health study over 11 yrs. Eur. Respir. J. 2005, 26, 45–51.
- Liu, C.-L.; Wu, C.-L.; Lu, Y.-T. Effects of Age on 1-Second Forced Expiratory Volume Response to Bronchodilation. Int. J. Gerontol. 2009, 3, 149–155.
- Kelly, R.S.; Sordillo, J.E.; Lutz, S.M.; Avila, L.; Soto-Quiros, M.; Celedón, J.C.; McGeachie, M.J.; Dahlin, A.; Tantisira, K.; Huang, M.; et al. Pharmacometabolomics of Bronchodilator Response in Asthma and the Role of Age-Metabolite Interactions. Metaboites 2019, 9, 179.
- Sordillo, J.E.; Lutz, S.M.; Kelly, R.S.; McGeachie, M.J.; Dahlin, A.; Tantisira, K.; Clish, C.; Lasky-Su, J.; Wu, A.C. Plasmalogens Mediate the Effect of Age on Bronchodilator Response in Individuals With Asthma. Front. Med. 2020, 7, 38.
- Kelly, R.S.; Sordillo, J.; Lasky-Su, J.; Dahlin, A.; Perng, W.; Rifas-Shiman, S.L.; Weiss, S.T.; Gold, D.R.; Litonjua, A.; Hivert, M.-F.; et al. Plasma metabolite profiles in children with current asthma. Clin. Exp. Allergy 2018, 48, 1297–1304.
- Van Der Sluijs, K.F.; Van De Pol, M.; Kulik, W.; Dijkhuis, A.; Smids, B.S.; Van Eijk, H.W.; Karlas, J.; Molenkamp, R.; Wolthers, K.C.; Johnston, S.; et al. Systemic tryptophan and kynurenine catabolite levels relate to severity of rhinovirus-induced asthma exacerbation: A prospective study with a parallel-group design. Thorax 2013, 68, 1122–1130.
- Collipp, P.J.; Chen, S.Y.; Sharma, R.K.; Balachandar, V.; Maddaiah, V.T. Tryptophane metabolism in bronchial asthma. Ann. Allergy 1975, 35, 153–158.
- Fogarty, A.; Broadfield, E.; Lewis, S.; Lawson, N.; Britton, J. Amino acids and asthma: A case-control study. Eur. Respir. J. 2004, 23, 565–568.




