
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Diego Sastre | + 6351 word(s) | 6351 | 2021-07-22 16:19:04 | | | |
| 2 | Conner Chen | Meta information modification | 6351 | 2021-08-02 14:29:40 | | |
Video Upload Options
The bacterial flagellum is a complex and dynamic nanomachine that propels bacteria through liquids. It consists of a basal body, a hook, and a long filament. The flagellar filament is composed of thousands of copies of the protein flagellin (FliC) arranged helically and ending with a filament cap composed of an oligomer of the protein FliD. The overall structure of the filament core is preserved across bacterial species, while the outer domains exhibit high variability, and in some cases are even completely absent. Flagellar assembly is a complex and energetically costly process triggered by environmental stimuli and, accordingly, highly regulated on transcriptional, translational and post-translational levels. Apart from its role in locomotion, the filament is critically important in several other aspects of bacterial survival, reproduction and pathogenicity, such as adhesion to surfaces, secretion of virulence factors and formation of biofilms. Additionally, due to its ability to provoke potent immune responses, flagellins have a role as adjuvants in vaccine development.
1. Flagellins
Flagellins are elongated proteins that constitute the flagellar filament and, apart from the FliD protein at the tip, are the only structural component of the filament. Their molecular weight ranges from 26 kDa in Bacillus cereus to 115 kDa in Desulfotalea psychrophila [1][2]. In E. coli and S. Typhimurium flagellins they are typically 51 kDa, although this varies among strains; for example the symbiotic strain E. coli Nissle 1917 has a flagellin protein of 61 kDa, while in some Salmonella mutants functional flagellins of 41 and 42 kDa have been reported [3][4]. Terminal regions of flagellins are highly homologous among species and they build the core of the filament [2]. Conversely, a hypervariable region in the middle of the polypeptide sequence makes outer domains significantly different, and in some bacteria such as B. subtilis it is completely absent [2][5].
The flagellin genes are known under various names depending on the species, such as fliC in E. coli, P. aeruginosa, and Salmonella, hag in B. subtilis, and flaA in C. jejuni and Helicobacter pylori. The number of flagellin genes also varies. Approximately 45% of bacteria have more than one flagellin-encoding gene in their genomes, the most extreme case being Magnetococus with 15 [6]. While the E. coli genome harbors only one flagellin gene, fliC, Salmonella has two, fliC and fljB [7]. However, only one of them is expressed at any point; the switch between them is called phase variation [8]. These two proteins diverge in the middle region providing distinct antigenicities. The studies on mutants locked in one of the two phases showed the different swimming properties and advantages of the FliC-expressing strains in colonization and infection of the gastrointestinal tract [9][10].
In other species with two flagellins such as H. pylori, Campylobacter spp. or Shewanella putrefaciens, both are present in the filament, the minor flagellin predominantly in the proximal region, while the major flagellin forms the remainder of the filament [11][12][13][14]. Although knocking out the minor flagellin does not usually affect the length of the filament, mutants exhibit changes in motility, especially in more viscous media, suggesting that having two flagellins in the filament provides optimal swimming characteristics in different environments [14][15]. The minor flagellin of C. jejuni also plays a role in defense against bacteriophage infection [16].
At the species level, P. aeruginosa has two types of flagellin genes, A and B, which differ in molecular weight and recognition by antibodies [17][18]. They are not, however, simultaneously present in the genome, but rather they are strain-specific with strains such as PAK containing type A FliC, while strains like PAO1 have type B FliC. While their N- and C-termini are highly conserved, the central region of the type A FliC is significantly shorter compared to the type B [19].
The number of flagellins in one cell can be as high as six (Caulobacter crescentus and Vibrio vulnificus) or even seven (Rhizobium leguminosarum) [6][20][21]. In V. vulnificus, mass spectrometry confirmed the presence of five distinct flagellins in the filament, with three having higher impact on motility, adhesion and cytotoxicity [20]. Some of these are redundant, such as for C. crescentus in which five of six flagellins can make filaments alone [6].
1.1. Flagellin Structure
Early biophysical characterization showed that the N- and C-termini of monomeric flagellin are mainly disordered and critical for polymerization, although they become highly structured in the filament [22][23]. Due to the tendency of full-length flagellin to polymerize, crystallization of S. Typhimurium flagellin FliC was achieved only after the removal of its terminal regions by limited proteolysis [24]. The overall shape of FliC obtained by combining X-ray crystallography and cryo-EM approaches revealed a structure in the form of a Greek letter Γ with four distinct domains (Figure 1a) [25][26]. Starting from the N-terminus, the polypeptide chain makes a single helix that forms one half of an α-helical domain called D0, continues with the partial formation of domains D1 and D2, and forms domain D3 prior to completing the folds of the previous three domains. Thus, domains D0-D2 have N- and C-terminal moieties.
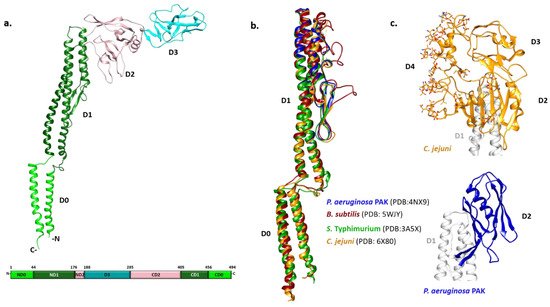
Figure 1. Structural characteristics of flagellins. (a) FliC monomer from S. Typhimurium with four distinct domain (PDB: 3A5X). Domains D0, D1 and D2 have N- and C-teminal moities, while D3 is formed by a continuous polypeptide sequence in the middle; (b) Structural alignment of D0 and D1 domains from various species showing high structural homology among them; (c) Outer domains of the C. jejuni flagellin FlaA and P. aeruginosa PAK type a FliC.
The N- and C-terminal helices of D0 form a coiled coil in the filament. The organization of D1 with three α-helices and a β-hairpin resembles a four-helix bundle with an extensive hydrophobic core. D2 and D3 consist primarily of β-strands organized in a specific fold called β-foliums in which the tips of the β-hairpins are bent or twisted [25][26].
While all flagellin structures known so far exhibit a high degree of structural similarity in the D0 and D1 region, the structures of the variable regions differ extensively (Figure 1b,c). The structure of the P. aeruginosa type A FliC lacking the D0 domain showed that the D1 domain is highly similar to its counterpart in S. Typhimurium [27]. Conversely, the D2 domain adopts a different fold with two β-sheets and one α-helix between them forming a less flexible cup-like structure positioned parallel to the D1 domain, instead of pointing away from it as in Salmonella (Figure 1c). In the case of Campylobacter, the variable region of the flagellin FlaA is larger and forms three domains (D2, D3 and D4) (Figure 1c) [28]. The D2 and D3 domains of the Campylobacter FlaA are structural homologs of each other and of the Pseudomonas FliC D2. D4 is inserted between C-terminal moieties of D2 and D1, with most of the FlaA glycans located on this domain. It is the most exposed outer domain resembling a shield for both D2 and D3 and, contrary to them, D4 does not have structural homologs.
1.2. Posttranslational Modifications of Flagellins
The presence of posttranslational modifications in flagellins was suspected early on, but their extent and importance became apparent only recently. Investigating discrepancies between theoretical and observed molecular weights showed that Pseudomonas type A flagellin was glycosylated [19]. A genomic island between flgL and fliC genes containing 14 open reading frames (ORFs) is responsible for glycosylation and it specifically modifies type A flagellin, while having no effect on type B [29]. These glycan chains in the PAK strain are O-linked to residues T189 and S260 through rhamnose and vary in length (up to 11 monosaccharides) and composition [30].
The type B flagellin characteristic of the PAO1 strain also contains O-linked glycans at serine residues 191 and 195, although in this case it is a much simpler modification with a single monosaccharide [31]. The genomic island responsible for type B glycosylation is smaller than its type A counterpart and contains four ORFs. Pseudomonas type A and B mutants in which glycosylation is absent or incomplete do not affect motility in general, although the lack of glycosylation significantly decreased virulence of both PAK and PAO1 strains in a burned-mouse model of infection [32]. Apart from O-linked glycans, N-linked glycosylation was also reported in P. aeruginosa strain PA14, with three sites on the D0 and D1 domains proposed as the most likely candidate positions [33].
Campylobacter flagellin is glycosylated at 19 serine and threonine residues with O-linked pseudaminic acid or related derivatives [34][35][36]. Glycosylation in C. jejuni is responsible for the 6 kDa difference between predicted and experimentally obtained molecular weight. Approximately 50 genes in a locus that is adjacent to the flaA and flaB genes encode the Campylobacter glycosylation machinery [37]. Glycosylation of flagellin in Campylobacter is necessary for filament assembly as mutants that produce unglycosylated flagellin do not have filaments and flagellin accumulates intracellularly [38].
H. pylori flagellins are also glycosylated with pseudaminic acid with most glycosylation sites in the central core region of the protein [39]. As in Campylobacter, glycosylation plays an important role during assembly of the filament; abolishing glycosylation leads to loss of the filament and motility, even though the levels of flagellin mRNA are unaffected [39].
The flagellin of Listeria monocytogenes has O-linked N-acetylglucosamines at up to six sites located in the central surface exposed region of the protein [30]. The flagellin of P. syringae is also glycosylated and these modifications are important for its virulence and swarming [40][41].
Another modification that is present in flagellins is methylation. In S. Typhimurium, the presence of methylated lysines was reported as early as 1959 [42]. Lysine methylation is performed by the enzyme FliB. Abolishing methylation does not affect filament assembly or motility [43]. However, it was shown that it promotes adhesion and host cell invasion [44]. N-methylation was also detected in Shewanella oneidensis [45].
2. Filament
The filament is a tubular structure made of more than 20,000 copies of the flagellin protein arranged in a helical fashion [46]. Flagellin is rotated and translated 11 times along the screw axis, making two turns, before it reaches its original position. Such an arrangement results in 11 stacks of flagellin along the axis called protofilaments, which are the basic functional units of the filament (Figure 2a). Depending on the interaction between flagellin molecules in a protofilament, it switches between two different states, left-handed (L) and right-handed (R) relative to the longitudinal axis, independently of its neighboring protofilament (Figure 2b) [47]. During swimming, native filaments adopt a superhelical or corkscrew shape and their rotation creates thrust. Depending on the number of L- and R-protofilaments, the pitch of the superhelix changes, modulating the swimming properties [48]. If all protofilaments are in the L- or R-state, the filament is straight and cannot create a thrust and, thus, cells are immotile. Apart from handedness, L- and R-filament also differ in length, with the R-filament being 1.5% shorter. During the normal smooth swimming of S. Typhimurium and E. coli, two R- and nine L-protofilaments form a filament that is a left-handed superhelix with a pitch of around 2–3 µm. The increased number of R-protofilaments eventually results in right-handed curly flagella with a shorter pitch typical for tumbling movements [48].
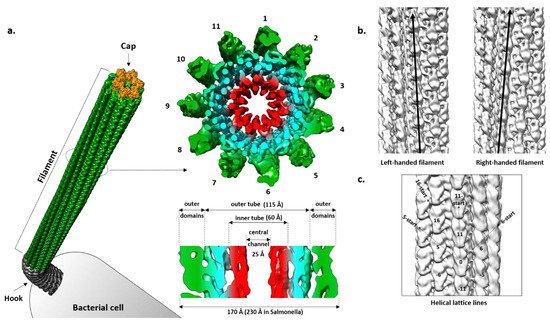
Figure 2. Cryo-EM structure of the flagellar filament from P. aeruginosa PAO1. (a) Cross-section of the left-handed filament consisting of 11 protofilaments. The inner tube (red) made of D0 domains and outer tube (cyan) together form the densely packed inner core of the filament. Outer domains are shown in green; (b) Left- and righ-handed filaments with all protofilaments tilted to the left and right from the central axis; (c) Helical symmetry lines in the filament. EMD: 8855, 8856.
Straight filaments in which all protofilaments are locked in either L- or R-state are much more amenable for structural studies than the native wavy forms in which, due to the presence of a mixture of L- and R-protofilaments, the strict helical symmetry is disturbed [49]. Several mutations of the fliC gene in Salmonella result in formation of straight filaments and were used in X-ray and EM studies [47]. Additionally, these mutations served as a basis to design the equivalent L- and R-mutants in other bacteria like P. aeruginosa and C. jejuni and to study their structures [28][50].
A cross-section of the Salmonella filament reveals a densely packed helical core 115 Å in diameter around a central channel that is ~25 Å wide, and an outer region exposed to the surface where domains of the individual molecules are clearly separated [51][52]. The diameter of the whole filament is ~230 Å. The core is organized into two tubes, inner and outer, connected through eleven short spokes, while the outer region contains two domains that are clearly defined.
Using cryo-EM, Yonekura et al. [26] obtained an electron density map of ~4 Å resolution that allowed them, in combination with the already known structure of FliC, to build the first complete atomic model of the R-filament from Salmonella. This model greatly contributed to understanding of the filament architecture, especially of its core, where the boundaries between domains of individual molecules were not obvious.
In regard to geometry, the Salmonella R-filament is a helical structure in which the FliC molecule is rotated 65.8° to the right followed by the translation of 4.7 Å. Protofilaments are tilted to the right by 3.5° relative to the longitudinal axis. The inner and outer tube are made of the D0 and D1 domains, respectively. In the inner tube, interactions between subunits are found along 11-start and 5-start helices and they are mostly hydrophobic, stabilizing the filament. The outer tube has a similar pattern with interactions found in the 11-start, 5-start and 16-start helices but they are predominantly polar–polar and charge–polar. Along the 11-start helix, the D1 domain of the upper subunit forms a concave surface that is complementary to the convex surface of the lower subunit and the two neighboring molecules make numerous Van der Waals contacts [25]. In the 5-start direction, the N-terminal helices of one molecule interact with the β-hairpin and C-terminal helix of the other molecule. N-terminal helices also make contact with the spoke in the 16-start direction. According to this rigid model, except for the small region of D2 proximal to the core, D2 and D3 do not engage in obvious inter-molecular contacts and they are projected away from the core. However, they do contribute to the overall stability of the filament, since deletion of the large part of this region makes the filament much more fragile in comparison to the wild type and mutant filaments, showing a contact between the adjacent outer domains along the 5-start helix that is absent in the mutant [4][53]. Therefore, it is likely that D2 and D3 exhibit flexibility in vivo, transiently interacting with the neighboring subunits and contributing to the filament integrity.
Recently, the L- and R-type filament structures of several other Gram-positive and Gram-negative bacteria became available, confirming that the 11-protofilament organization is common for all species and that the core is structurally conserved. While B. subtilis does not have any outer domains, D2 and D3 in P. aeruginosa appear as a single domain in which the D3 portion extends along the axis and seems to form a dimer with the D2 of the subunit above, although the structure could not be resolved due to low resolution in that region [50]. Another distinctive feature of the Pseudomonas filament is the existence of a seam along one of the protofilaments, which introduces a non-helical perturbation. This perturbation is characteristic of the so-called complex filaments that are found in some species like Rhizobium lupini or Pseudomonas rhodos, but in P. aeruginosa it is present only at the level of the outer domains [50]. As mentioned previously, the outer region in Campylobacter is significantly larger containing 3 domains and, apart from interactions along the protofilament, it also makes contacts in 5-start and 6-start directions, contributing to filament integrity [28].
2.1. Comparison of L- and R-Filaments
One of the questions that draws much attention is the structural basis for the polymorphic switching of the filament. Comparison of the L- and R-structures obtained by cryo-EM revealed that the D0 domains align well with some local changes in conformation, while the rest of the molecule is twisted by 5° in the counterclockwise direction [54]. Previous X-ray diffraction study on L- and R-types from Salmonella showed that there were no changes in helical parameters in the inner region of the core made of D0 domains [55]. However, the two types differ in the outer regions and the distance between two subunits in the protofilament is 0.8 Å shorter in the R-type compared to the L-type. Maki-Yonekura et al. [54] found that the packing of the hydrophobic side chains at the lower portion of D1 is different between L- and R-type and that the intersubunit interactions along the 5-start helix are important in stabilizing the conformation. Based on molecular dynamics simulations of the polymorphic supercoiling mechanism, Kitao et al. [56] identified three types of interaction between subunits: permanent (the same pair of residues in different states), sliding (the same types of interaction with variable partners) and switch interactions that were responsible for locking the protofilament interface in R- or L-handed state. The switch interactions were found only along the 5-start helices. However, this was not supported by the analysis of the corresponding interactions in L- and R-filaments of B. subtilis [50].
2.2. Filament Cap
Originally designated as a HAP2, FliD is localized at the tip of the filament where several copies of it assemble into a capping structure. The presence of the cap prevents the leakage of flagellin that is continuously exported through the flagellar type III secretion system into the filament channel and facilitates its insertion at the growing end of the filament. Early cryo-EM studies of the Salmonella cap showed that it is a stool-like pentameric structure with the flat star-shaped head region and five legs positioned almost at the right angle. The head region is 145 Å in dimeter and the height of the cap is 125 Å.
While they vary in size, FliDs from different bacteria have conserved N- and C-terminal regions predicted to be coiled-coils and are largely unstructured in the monomeric state [57]. The first high-resolution structure of the P. aeruginosa FliD showed that this 50 kDa protein has three distinct domains: a highly flexible D1 domain that corresponds to the leg domains seen in the cap structure, and compact D2 and D3 domains that comprise the head [58]. Much as in flagellin, a continuous sequence in the middle of the polypeptide folds into D3 while domains D1 and D2 consist of N- and C-terminal portions of the chain relative to D3. Domains D2 and D3 are each made of two antiparallel β-sheets, while D1 could not be resolved with confidence apart from one helix. The structure of the E. coli FliD offered more insight into D1, showing that it is a helical bundle of at least four helices with a β-hairpin similar to the flagellin D1 domain, although approximately 50 terminal residues from both sides were not present in the construct (Figure 3a,c) [59]. These structures, together with the ones from Salmonella and Serratia marcescens [59][60], indicated that the structural organization of the capping proteins among different bacteria is preserved. However, not all FliDs are of the same size. For example, the FliD of H. pylori is 74 kDa and the crystal structure revealed that apart from domains D1–3 present in the previously studied proteins, it has two additional domains, D4 and D5, both of which reside in the head region with similar overall folds to D2 and D3 (Figure 3b) [61]. These domains may be related to the specific role FliD plays as a virulence factor during H. pylori infection; stronger antigenicity of D4 and D5 further supports this [61].
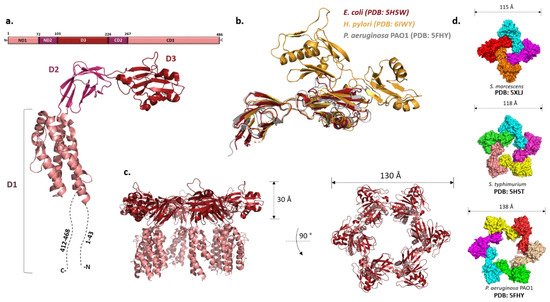
Figure 3. FliD and the filament cap. (a) Structure of FliD from E. coli shows three domains (D1–D3) (PDB: 5H5V). The N- and C-termini are highly flexible and could not be resolved. (b) Structural alignment of FliD molecules from different species show high structural homology of D2 and D3 domains. FliD from H. pylori has two additional domains, D4 and D5. (c) Crystal structure of the filament cap from E. coli showing a hexameric structure. Domains D2 and D3 form the head region, while D1 (incomplete) forms the flexible leg region; (d) Oligomeric states of the crystallized FliD caps differ across species.
Once the hook and the junction are formed, FliD forms the filament cap, promoting the polymerization of FliC. The oligomeric state of the filament cap is still an open question. Initial characterization of the recombinant FliD of Salmonella showed the existence of decamers in solution, but the combination of cross-linking experiments, analytical ultracentrifugation and electron microscopy revealed that a pentamer is the functional unit, decamers being the product of a non-physiological cap dimerization. This was generally also accepted as the case for other bacteria, and the recent cryo-EM structure of the pentameric C. jejuni cap supports it [62]. However, both Pseudomonas and E. coli FliDs crystallized as hexamers and Serratia FliD as a tetramer (Figure 3d) [58][59][60]. Moreover, tetramers and pentamers of Pseudomonas FliD and tetramers of Salmonella FliD were observed in vitro [58][62]. While these stoichiometries could arise due to non-physiological conditions, in the case of P. aeruginosa hexamers were also detected in vivo [58].
The mechanism of cap-filament interaction is yet to be explained. During filament elongation, new flagellin molecules arrive at the tip continuously. This would require high mobility of the cap and continuous conformational changes of the D1 domains, most probably independently from one another, in order to accommodate arriving FliC subunits, all while staying firmly attached to the filament. One way in which FliD could interact with FliC is through its highly flexible terminal regions, present in all proteins in the outer part of the flagellum and are shown to mediate protein–protein interactions.
3. Protein Synthesis, Export and Filament Elongation
3.1. Genetic Regulation of FliC and FliD
The genetic organization of the flagellar components is highly complex and relatively well conserved across bacteria with the same type of flagellar arrangement (i.e., peritrhichous, monotrichous/polar, etc.) but quite different between them [63][64][65]. The bacterial flagellar regulon commonly consists of dozens of genes grouped in several operons, encoding all structural proteins of the flagellum, the chemosensory apparatus and the regulators that control gene expression. For example, there are about 50 genes involved in the synthesis of the polar P. aeruginosa flagella, distributed in 17 putative operons comprising 41 flagellar genes (26 encoding structural proteins, eight encoding regulators and six involved in the export apparatus), linked to nine genes involved in chemotaxis and clustered in three regions of the chromosome constituting the fla regulon [66][67]. On the other hand, in peritrichous bacteria such as Salmonella spp. and Escherichia coli, the flagellar regulon contains 70 genes distributed in at least 25 operons [43][64]. The transcription of these genes is hierarchical and controlled by different promoter classes that are temporally regulated (Figure 4, Table 1). In peritrichous bacteria, flagellar genes are organized into three classes (I-III), starting with the transcription of the class I master operon as a response to the environmental stimuli that leads to the synthesis of the transcriptional regulator FlhDC. This regulator, in turn, activates the transcription of the class II genes responsible for the assembly of the hook-basal body (HBB) complex. Among class II genes are two crucial regulators of the class III genes: transcription factor FliA (σ28) responsible for the transcription of class III genes, and FlgM, which acts as a FliA inhibitor. Once the HBB is assembled, FlgM is exported out of the cell via the HBB central channel, releasing FliA to interact with RNA polymerase and guide the transcription of the class III genes, including flagellin. This three-tiered organization ensures that cells produce flagella in response to the environmental signals and that energetically costly synthesis of the filament proceeds only when enough HBBs have been formed. An exception is the Actinoplanes missouriensis zoospore in which there is no hierarchical coordination and 33 flagellar genes are transcribed simultaneously during the sporangium formation [68]. The filament cap gene fliD belongs to the group of class II genes, whereas flagellin genes belong to class III. Although fliD is expressed as a class II gene, it is not assembled into the growing flagellar structure until the hook and the junction proteins are expressed from class III (including flagellin genes) and incorporated into the flagellar organelle [69]. Hence, this cascade serves to control the timing of gene expression to coincide with the assembly of the flagellar apparatus and filament.
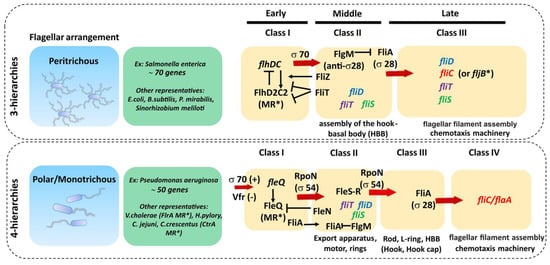
Figure 4. Genetic organization of main flagellar proteins involved in filament formation. The transcription of flagellar genes is hierarchical and controlled by different promoter classes that are temporally regulated. In peritrichous bacteria, flagellar genes are organized into three classes (I–III) and in monotrichous/polar bacteria the flagellar genes are organized into four classes, both starting with the transcription of the class I master regulator (MR*). Red arrows indicate the proteins and/or transcription factors responsible for the transcription activation of each corresponding class of flagellar genes. Black narrow arrows indicate positive activation/induction and black arrows with a vertical line tip (|) indicate inhibition. Genes involved in filament synthesis and their cognate chaperones are represented in italic colored font.
Table 1. Flagellins and FliD and their regulators.
| Protein | Function | Sigma Factor | Class in Flagellar Regulatory Hierarchy | Transcriptional Expression Regulators | Post-Transcriptional Regulators | Specific Secretion Chaperone | |
|---|---|---|---|---|---|---|---|
| 3-Tiered | 4-Tiered | ||||||
| Flagellin (FliC, FlaA) | Structural component of the filament | Sigma28 Sigma54 Sigma43 |
Class III gene | Class IV gene | CodY Environmental factors (nutrients, c-di-GMP, ppGpp, BCAA, temperature…) |
Self-regulating CsrA-FliW |
FliS |
| FliD | Filament cap | Sigma70 Sigma28 Sigma54 |
Class II and III | Class II | Cognate flagellin | ? | FliT |
As mentioned above, S. Typhimurium exhibits phase variation, or alternate expression of two flagellin genes, fliC and fljB. This process is regulated by a control region upstream of fljB that is a subject of inversion, which orients the region forward and generates a promoter. This allows co-transcription of fljB and fljA, a transcriptional repressor that inhibits the expression of FliC [70].
Recently, Rao et al. found that flagellar gene expression in Salmonella is bimodal, meaning that in a population of genetically identical cells under the same conditions only a fraction of them is motile [71][72]. This bimodality is present at both the class II and class III levels and is governed by separate mechanisms. While a double negative feedback loop involving two flagellar regulatory proteins, RflP and FliZ, controls the expression of the class II genes in response to nutrient availability, class III gene expression is tuned by the secretion of FlgM and there is a minimum number of HBBs necessary for the cell to pass this checkpoint.
In bacteria with a polar flagellum, flagellar genes are transcribed in a four-tiered hierarchy, with genes encoding components of the HBB split between class II and class III (Figure 4) [66]. Gene regulation in Pseudomonas is more complex and involves the activation of the two-component system FleS-FleR and another sigma factor RpoN (σ54) to activate class III genes. In addition, unlike in Salmonella, FliA (σ28) is constitutively expressed in Pseudomonas independently of other flagellar genes, which makes it one of the class I genes [66].
In bacteria with multiple flagellins, such as Helicobacter and Campylobacter, expression is under the control of different sigma factors with FlaA under control of σ28, while FlaB is under control of σ54 [73][74]. Although the major flagellin is usually under the control of σ28, there are exceptions such as V. cholera and S. oneidensis in which the major flagellin is dependent on σ54, while commensal gut bacteria of Eubacterium and Roseburia species are under the control of σ28 and σ43 [75][76]. In the phylogenetically related Butyrivibrio fibrisolvens, transcription of one fliC gene is driven from two different promoters, yielding two transcripts with alternative transcription start-sites [76].
In some Gram-positive bacteria, changes in the environmental conditions such as nutrient limitation induce variations in the levels of the intracellular messengers guanosine tetra/pentaphosphate, (p)ppGpp, guanosine nucleoside triphosphate (GTP) and branched chain amino-acid pools. These variations are detected by a conserved GTP-sensing protein CodY and a global regulator CsrA that modulate flagellin expression. Cell motility can be repressed under elevated intracellular cyclic-di-GMP levels, by impeding transcription of some of the flagellum genes [77][78][79][80][81]. Indeed, the expression of fliD and fliC is repressed in phosphodiesterase 3 (PDE3) knockout mutants triggered by elevated c-di-GMP accumulation [82].
Because the presence of flagellin can be deleterious to the bacterium on account of inducing host immunity, flagellated bacteria downregulate (or turn off) flagellin expression during the host invasion to avoid host immune responses. Therefore, normal microbiota within the healthy adult mammalian gut has been shown to have overall relatively low levels of flagellin expression, while TLR5−/− mice exhibited a diversity of gut microbiome members with overexpressed flagellar genes [83][84].
Both commensal gut strains of motile E. coli and pathogenic S. Typhimurium strains strongly down-regulate their genes coding for flagellar machinery and lose their motility once inside the host [85][86]. Under environmental temperature conditions (22–30 °C), the expression of flagellar genes in the human pathogens Listeria monocytogenes and Legionela pneumophila is normal, but significantly reduced when raised to 37 °C, revealing temperature-dependent transcription mainly controlled by the protein GmaR, which acts as a protein expression thermostat [87][88]. These types of system provide a pathogen with the ability to turn off immune-stimulating antigens before they trigger adverse host defense mechanisms once inside their target host.
Regulation of flagellin expression also occurs on the post-transcriptional level by proteins that bind to the untranslated leader region of the flagellin mRNA, affecting transcript stability and/or ribosome access [89][90][91]. For instance, the RNA-binding protein CsrA of B. burgdorferi specifically mediates synthesis of the major flagellin by inhibiting translation initiation of its transcript. In some cases, post-transcriptional regulators repress the accumulation of flagellin when cells are defective for flagellar hook assembly [92][93].
An assembly checkpoint that prevents flagellin translation and assembly prior to hook completion was discovered in B. subtilis. This is governed by a homeostatic mechanism in which the flagellin protein itself is a critical regulator. In a partner switching mechanism, the flagellar assembly factor FliW binds flagellin, while a global regulator CsrA binds flagellin mRNA, repressing its translation. After completion of the hook, flagellin is secreted, and the released FliW binds CsrA, thus derepressing flagellin translation [94]. An interesting case is A. missouriensis, in which all flagellar genes are transcribed simultaneously during sporangium formation and there is no checkpoint mechanism in the process of flagellar gene transcription to optimize the efficiency of the flagellar assembly [68]. The post-transcriptional level of regulation between protein production and assembly could play an important role, although this remains unknown.
3.2. Chaperones
Accumulation and premature oligomerization of flagellar axial proteins in the cytosol could be wasteful and detrimental to the cell. For this reason, bacteria encode flagellar-specific chaperones dedicated to facilitating the assembly and export of the flagellum components mainly by protecting and/or preventing their cognate flagellar protein substrates from aggregation or avoiding premature undesired interactions of flagellar proteins in the cytoplasm prior to interaction with the export gate [95]. Due to the small diameter of the flagellar export central channel (20–30 Å), the proteins destined for incorporation into the growing flagellum must be exported in a partially or completely unfolded state, implying that premature folding and oligomerization in the cytosol must be prevented to keep them in a secretion-competent conformation for flagellum assembly [96]. Hence, the external flagellar components require T3SS-specific chaperones to facilitate their efficient export [64].
3.2.1. FliS—Flagellin Chaperone
FliS acts as a flagellin-specific T3SS chaperone of FliC, preventing premature folding and inappropriate interaction of newly synthesized flagellin subunits in the cytosol, thus facilitating its export and polymerization upon completion of the HBB assembly. Yeast two-hybrid assays indicated that the C-terminal disordered region of flagellin is essential for FliS binding and, accordingly, spontaneous mutations causing flagellin accumulation in the cytoplasm map to the C-terminal region of FliC [97]. Thermodynamic experiments indicated that FliS does not function as an anti-folding factor keeping flagellin in a secretion-competent conformation. Instead, FliS binding stabilizes the flagellin conformation through formation of the α-helical secondary structure in the last 40 C-terminal residues of FliC (residues 454–494) [70][96][98].
The first crystal structure of a flagellar chaperone, Aquifex aeolicus FliS (AaFliS), revealed a novel, mainly α-helical fold, different from those of the T3SS chaperones [99]. The structure of AaFliS chaperone in complex with a C-terminal fragment of its cognate flagellin (PDB: 1ORY) shows a monomer-to-monomer interaction. However, it was also reported that FliS binds in a 2:1 stoichiometry to the C-terminal region of flagellin to completely prevent the premature polymerization of newly synthesized flagellin molecules, as a 1:1 FliS to FliC molar ratio was able to prevent FliC polymerization only partially. Intriguingly, no stable FliS–FliC complex could be detected in S. typhimurium cell extracts subjected to gel-filtration chromatography, suggesting that this interaction may be transient in vivo. Such rapid chaperone dissociation would favor the subsequent export of FliC [100].
The interaction between FliS chaperone and FlhA is a key step preceding the efficient transfer of FliC to the platform of the flagellar type III export apparatus for a rapid export during flagellar filament assembly [101][102][103]. Filament proteins in complex with their cognate chaperones bind to a highly conserved hydrophobic pocket of FlhAC to promote unfolding and protein translocation by the protein export apparatus. It was predicted that different binding affinities of FlhAC for the chaperone substrate complexes may be the key to defining the correct order of protein export among the filament-type proteins [104]. Recently, a structure of the ternary complex formed by FliC, FliS and the export gate protein FlhA revealed that FliC does not interact directly with FlhA (Figure 5). Instead, the presence of FliC induces a binding-competent conformation of FliS that exposes the motif which is specifically recognized by FlhA [105]. Moreover, SAXS and HDX-MS experiments showed the formation of a heterotrimeric FliC-FliS-FliW complex that interacts with FlhA suggesting that FliS and FliW are released during flagellin export. FliW and FliS bind to opposing interfaces located at the N- and C-termini of flagellin, respectively, and these proteins seems to synchronize the production of flagellin with the capacity of the T3SS to secrete flagellin [106].
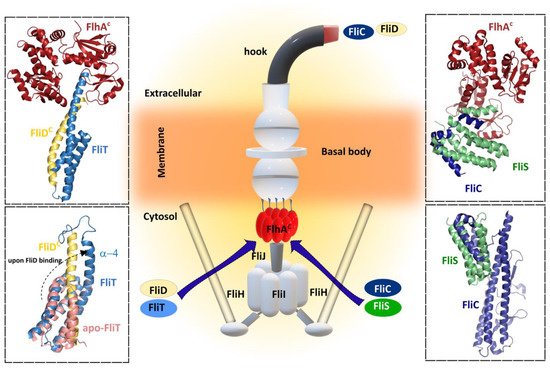
Figure 5. Bacterial flagellar filament proteins and their cognate specific chaperone form complexes with FlhA for filament exportation/synthesis. FliC and FliD proteins interact with each cognate chaperone FliS and FliT, respectively, before interacting with the cytosolic region of FlhA (FlhAC). Upper left—Cartoon representation of crystal structure 6CH2 (FlhAC-FliT-FliD complex from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2. Upper right—Cartoon representation of crystal structure 6CH3 (FlhAC-FliS-FliC complex) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2). Lower left—Superposition of cartoon representation of both NMR solution structures: 5KRW (FliD-FliT) and 5KS6 (apo-FliT) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2. The FliT helix α4 is released upon FliD binding. Lower right—Cartoon representation of crystal structure 5MAW (FliS-FliC complex) from B. subtilis. Center figure—simplified schematic representation of the main flagellar components.
FliS plays an additional role in suppressing the secretion of FlgM in Salmonella. The loss of FliS results in a short filament phenotype despite high expression levels of FliC, which is explained by the increase in the secretion level of FlgM [101]. Bypass mutants have been isolated from a Salmonella ΔfliS mutant, and all those mutations were identified in FliC [107][108].
Galeva et al. and Xu et al. investigated the direct interaction between FliS and FlgM from S. typhimurium and Yersinia pseudotuberculosis, respectively [100][109]. Using a number of different approaches, they showed that these proteins specifically interact to form a 1:1 complex, and that this interaction protects FlgM from proteolysis. FliS acts as a negative inhibitor of FlgM secretion, keeping this intrinsically disordered protein stable before FliA is expressed in cells. The binding site of FliA on FlgM is close to or even overlaps with the binding site of FliS, suggesting that FliA binding removes FliS from the complex. In addition, FliS from S. typhimurium is expressed from both class II and class III promoters, while FliC is expressed only from a class III promoter, indicating that FliS is synthesized and stabilized by FlgM prior to FliC (or FljB) production.
3.2.2. FliT—Chaperone of FliD
For the correct formation of the filament, a filament-capping protein FliD should be exported first during the filament assembly to form a penta- or hexameric cap that promotes self-assembly of FliC. In order to do this, FliD requires the assistance of its chaperon FliT [110]. FliT act as key flagellar chaperone in the assembly and operation of the flagellum because it binds to several flagellar proteins in the cytoplasm, such as the export apparatus components FliI, FliJ, and FlhA, beyond interaction with its cognate FliD. As an example of its versatility, FliT also functions as a negative transcriptional regulator of flagellar genes by inhibiting the formation of a DNA complex with the master regulator FlhDC. Lately, several crystal structures of FliT alone and in complex with FliD or FliI have become available [95][110]. Although in the crystal structure of Salmonella FliT was present as a tetramer, the observation that FliT was in the monomer–dimer equilibrium under physiological conditions suggests that the tetrameric form was an artefact of crystal packing. The structure revealed, however, that FliT adopts an antiparallel four α-helix bundle and uses a hydrophobic surface formed by the first three helices to recognize its substrate proteins. In the absence of a substrate protein, FliT adopts an auto-inhibited structure conformation in which both the binding site for the partner proteins (the hydrophobic surface formed by helices α1–α3) and the binding site for FlhA (helix α4) are occluded. This auto-inhibited structure may serve to protect the substate-binding surface from aggregation in the absence of substrates and to mask the binding site for FlhA, adopting an uninhibited conformation only when the substrate protein is to be targeted to the export gate. The auto-inhibition/activation and targeting mechanism reported for FliT appears to be shared among other flagellar chaperones such as FliS, which also adopts an auto-inhibited structure, released upon FliC binding. The N-terminus of FliS serves as the FlhA binding site, and in these chaperones a highly conserved Tyr residue appears to be essential for efficient FlhA binding, suggesting that these chaperones may use similar strategies for substrate binding and activation of the complexes for binding to FlhA and thus for targeting to the export gate. A recent structure of the FlhA−FliT−FliD ternary complex revealed that there is no direct interaction between FliD and FlhA, as was observed with FlhA–FliS–FliC ternary complex (Figure 5) [105].
Although the structural architecture of FliT is similar to that of FliS, the arrangement of α-helices is different. FliS forms a heterodimer with its cognate substrate FliC. The C-terminal region of FliC interacts with all three α-helices of FliS in an extended conformation. Similarly, FliT forms a stable heterodimeric complex with FliD in solution through an interaction between the C-terminal half of FliT and the C-terminal region of FliD, similar to that of the FliS–FliC complex. However, only the highly conserved surface-exposed Lys79 in α3, is critical for the interaction with FliD suggesting that the C-terminal region of FliD might interact with α3 of FliT in a different manner from the FliS–FliC interaction. Database searches revealed that FliS is the most widely conserved and ancient flagellar chaperone and it has been proposed that other flagellar chaperones, such as FlgN and FliT, has evolved from FliS. Biochemical and structural analyses have provided insight into how the C-terminus of FliT regulates its interactions with the FlhDC complex, FliI ATPase, and FliJ (subunits of the export apparatus that may function as a general chaperone), and the conformational change that is responsible for the switch between binding partners during flagellar protein export. After completion of HBB assembly, the C-terminal α4 helix of FliT is released from the hydrophobic cleft formed by the α2 and α3 helices to allow the FliT–FliD complex to bind to the FliH–FliI–FliJ complex through the specific interactions of FliT with FliI and FliJ, and the entire complex binds to the docking platform of the export gate. After FliD is unfolded and translocated into the flagellar channel to assemble the filament-capping structure at the tip, unbound FliT is free to interact with FlhC and form a complex FliT–FlhD4C2, thereby suppressing the class II gene expression, functioning as an anti-FlhD4C2 factor. FliT later dissociates from the FlhDC complex, allowing the free FlhDC complex to activate the transcription from the class II promoters anew.
References
- Il Kim, M.; Lee, C.; Park, J.; Jeon, B.Y.; Hong, M. Crystal structure of Bacillus cereus flagellin and structure-guided fusion-protein designs. Sci. Rep. 2018, 8, 5814.
- Beatson, S.A.; Minamino, T.; Pallen, M.J. Variation in bacterial flagellins: From sequence to structure. Trends Microbiol. 2006, 14, 151–155.
- Steimle, A.; Menz, S.; Bender, A.; Ball, B.; Weber, A.N.R.; Hagemann, T.; Lange, A.; Maerz, J.K.; Parusel, R.; Michaelis, L.; et al. Flagellin hypervariable region determines symbiotic properties of commensal Escherichia coli strains. PLoS Biol. 2019, 17, e3000334.
- Yoshioka, K.; Aizawa, S.; Yamaguchi, S. Flagellar filament structure and cell motility of Salmonella typhimurium mutants lacking part of the outer domain of flagellin. J. Bacteriol. 1995, 177, 1090–1093.
- LaVallie, E.R.; Stahl, M.L. Cloning of the flagellin gene from Bacillus subtilis and complementation studies of an in vitro-derived deletion mutation. J. Bacteriol. 1989, 171, 3085–3094.
- Faulds-Pain, A.; Birchall, C.; Aldridge, C.; Smith, W.D.; Grimaldi, G.; Nakamura, S.; Miyata, T.; Gray, J.; Li, G.; Tang, J.X.; et al. Flagellin redundancy in Caulobacter crescentus and its implications for flagellar filament assembly. J. Bacteriol. 2011, 193, 2695–2707.
- Macnab, R.M. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 1992, 26, 131–158.
- Silverman, M.; Zieg, J.; Simon, M. Flagellar-phase variation: Isolation of the rh1 gene. J. Bacteriol. 1979, 137, 517–523.
- Ikeda, J.S.; Schmitt, C.K.; Darnell, S.C.; Watson, P.R.; Bispham, J.; Wallis, T.S.; Weinstein, D.L.; Metcalf, E.S.; Adams, P.; O’Connor, C.D.; et al. Flagellar phase variation of Salmonella enterica serovar Typhimurium contributes to virulence in the murine typhoid infection model but does not influence Salmonella-induced enteropathogenesis. Infect. Immun. 2001, 69, 3021–3030.
- Horstmann, J.A.; Zschieschang, E.; Truschel, T.; de Diego, J.; Lunelli, M.; Rohde, M.; May, T.; Strowig, T.; Stradal, T.; Kolbe, M.; et al. Flagellin phase-dependent swimming on epithelial cell surfaces contributes to productive Salmonella gut colonisation. Cell Microbiol. 2017, 19, e12739.
- Kostrzynska, M.; Betts, J.D.; Austin, J.W.; Trust, T.J. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J. Bacteriol. 1991, 173, 937–946.
- Nuijten, P.J.; van Asten, F.J.; Gaastra, W.; van der Zeijst, B.A. Structural and functional analysis of two Campylobacter jejuni flagellin genes. J. Biol. Chem. 1990, 265, 17798–17804.
- Wassenaar, T.M.; Bleumink-Pluym, N.M.; Newell, D.G.; Nuijten, P.J.; van der Zeijst, B.A. Differential flagellin expression in a flaA flaB+ mutant of Campylobacter jejuni. Infect. Immun. 1994, 62, 3901–3906.
- Kuhn, M.J.; Schmidt, F.K.; Farthing, N.E.; Rossmann, F.M.; Helm, B.; Wilson, L.G.; Eckhardt, B.; Thormann, K.M. Spatial arrangement of several flagellins within bacterial flagella improves motility in different environments. Nat. Commun. 2018, 9, 5369.
- Guerry, P.; Alm, R.A.; Power, M.E.; Logan, S.M.; Trust, T.J. Role of two flagellin genes in Campylobacter motility. J. Bacteriol. 1991, 173, 4757–4764.
- Lis, L.; Connerton, I.F. The Minor Flagellin of Campylobacter jejuni (FlaB) Confers Defensive Properties against Bacteriophage Infection. Front. Microbiol. 2016, 7, 1908.
- Allison, J.S.; Dawson, M.; Drake, D.; Montie, T.C. Electrophoretic separation and molecular weight characterization of Pseudomonas aeruginosa H-antigen flagellins. Infect. Immun. 1985, 49, 770–774.
- Morgan, J.A.; Bellingham, N.F.; Winstanley, C.; Ousley, M.A.; Hart, C.A.; Saunders, J.R. Comparison of flagellin genes from clinical and environmental Pseudomonas aeruginosa isolates. Appl. Environ. Microbiol. 1999, 65, 1175–1179.
- Brimer, C.D.; Montie, T.C. Cloning and comparison of fliC genes and identification of glycosylation in the flagellin of Pseudomonas aeruginosa a-type strains. J. Bacteriol. 1998, 180, 3209–3217.
- Kim, S.Y.; Thanh, X.T.; Jeong, K.; Kim, S.B.; Pan, S.O.; Jung, C.H.; Hong, S.H.; Lee, S.E.; Rhee, J.H. Contribution of six flagellin genes to the flagellum biogenesis of Vibrio vulnificus and in vivo invasion. Infect. Immun. 2014, 82, 29–42.
- Tambalo, D.D.; Bustard, D.E.; Del Bel, K.L.; Koval, S.F.; Khan, M.F.; Hynes, M.F. Characterization and functional analysis of seven flagellin genes in Rhizobium leguminosarum bv. viciae. Characterization of R. leguminosarum flagellins. BMC Microbiol. 2010, 10, 219.
- Vonderviszt, F.; Kanto, S.; Aizawa, S.-I.; Namba, K. Terminal regions of flagellin are disordered in solution. J. Mol. Biol. 1989, 209, 127–133.
- Namba, K.; Yamashita, I.; Vonderviszt, F. Structure of the core and central channel of bacterial flagella. Nature 1989, 342, 648–654.
- Samatey, F.A.; Imada, K.; Vonderviszt, F.; Shirakihara, Y.; Namba, K. Crystallization of the F41 fragment of flagellin and data collection from extremely thin crystals. J. Struct. Biol. 2000, 132, 106–111.
- Samatey, F.A.; Imada, K.; Nagashima, S.; Vonderviszt, F.; Kumasaka, T.; Yamamoto, M.; Namba, K. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 2001, 410, 331–337.
- Yonekura, K.; Maki-Yonekura, S.; Namba, K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 2003, 424, 643–650.
- Song, W.S.; Yoon, S.I. Crystal structure of FliC flagellin from Pseudomonas aeruginosa and its implication in TLR5 binding and formation of the flagellar filament. Biochem. Biophys. Res. Commun. 2014, 444, 109–115.
- Kreutzberger, M.A.B.; Ewing, C.; Poly, F.; Wang, F.; Egelman, E.H. Atomic structure of the Campylobacter jejuni flagellar filament reveals how epsilon Proteobacteria escaped Toll-like receptor 5 surveillance. Proc. Natl. Acad. Sci. USA 2020, 117, 16985–16991.
- Arora, S.K.; Bangera, M.; Lory, S.; Ramphal, R. A genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc. Natl. Acad. Sci. USA 2001, 98, 9342–9347.
- Schirm, M.; Kalmokoff, M.; Aubry, A.; Thibault, P.; Sandoz, M.; Logan, S.M. Flagellin from Listeria monocytogenes is glycosylated with beta-O-linked N-acetylglucosamine. J. Bacteriol. 2004, 186, 6721–6727.
- Verma, A.; Schirm, M.; Arora, S.K.; Thibault, P.; Logan, S.M.; Ramphal, R. Glycosylation of b-Type flagellin of Pseudomonas aeruginosa: Structural and genetic basis. J. Bacteriol. 2006, 188, 4395–4403.
- Arora, S.K.; Neely, A.N.; Blair, B.; Lory, S.; Ramphal, R. Role of motility and flagellin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect. Immun. 2005, 73, 4395–4398.
- Khemiri, A.; Naudin, B.; Franck, X.; Song, P.C.; Jouenne, T.; Cosette, P. N-glycosidase treatment with 18O labeling and de novo sequencing argues for flagellin FliC glycopolymorphism in Pseudomonas aeruginosa. Anal. Bioanal. Chem. 2013, 405, 9835–9842.
- Doig, P.; Kinsella, N.; Guerry, P.; Trust, T.J. Characterization of a post-translational modification of Campylobacter flagellin: Identification of a sero-specific glycosyl moiety. Mol. Microbiol. 1996, 19, 379–387.
- Thibault, P.; Logan, S.M.; Kelly, J.F.; Brisson, J.R.; Ewing, C.P.; Trust, T.J.; Guerry, P. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 2001, 276, 34862–34870.
- Logan, S.M.; Kelly, J.F.; Thibault, P.; Ewing, C.P.; Guerry, P. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 2002, 46, 587–597.
- Szymanski, C.M.; Logan, S.M.; Linton, D.; Wren, B.W. Campylobacter—A tale of two protein glycosylation systems. Trends Microbiol. 2003, 11, 233–238.
- Goon, S.; Kelly, J.F.; Logan, S.M.; Ewing, C.P.; Guerry, P. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 2003, 50, 659–671.
- Schirm, M.; Soo, E.C.; Aubry, A.J.; Austin, J.; Thibault, P.; Logan, S.M. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 2003, 48, 1579–1592.
- Taguchi, F.; Shibata, S.; Suzuki, T.; Ogawa, Y.; Aizawa, S.; Takeuchi, K.; Ichinose, Y. Effects of glycosylation on swimming ability and flagellar polymorphic transformation in Pseudomonas syringae pv. tabaci 6605. J. Bacteriol. 2008, 190, 764–768.
- Chiku, K.; Yamamoto, M.; Ohnishi-Kameyama, M.; Ishii, T.; Yoshida, M.; Taguchi, F.; Ichinose, Y.; Ono, H. Comparative analysis of flagellin glycans among pathovars of phytopathogenic Pseudomonas syringae. Carbohydr. Res. 2013, 375, 100–104.
- Ambler, R.P.; Rees, M.W. Epsilon-N-Methyl-lysine in bacterial flagellar protein. Nature 1959, 184, 56–57.
- Frye, J.; Karlinsey, J.E.; Felise, H.R.; Marzolf, B.; Dowidar, N.; McClelland, M.; Hughes, K.T. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2006, 188, 2233–2243.
- Horstmann, J.A.; Lunelli, M.; Cazzola, H.; Heidemann, J.; Kuhne, C.; Steffen, P.; Szefs, S.; Rossi, C.; Lokareddy, R.K.; Wang, C.; et al. Methylation of Salmonella Typhimurium flagella promotes bacterial adhesion and host cell invasion. Nat. Commun. 2020, 11, 2013.
- Sun, L.; Jin, M.; Ding, W.; Yuan, J.; Kelly, J.; Gao, H. Posttranslational modification of flagellin FlaB in Shewanella oneidensis. J. Bacteriol. 2013, 195, 2550–2561.
- Yonekura, K. Growth mechanism of the bacterial flagellar filament. Res. Microbiol. 2002, 153, 191–197.
- Kamiya, R.; Asakura, S.; Yamaguchi, S. Formation of helical filaments by copolymerization of two types of ‘straight’ flagellins. Nature 1980, 286, 628–630.
- Calladine, C.R.; Luisi, B.F.; Pratap, J.V. A “mechanistic” explanation of the multiple helical forms adopted by bacterial flagellar filaments. J. Mol. Biol. 2013, 425, 914–928.
- Trachtenberg, S.; DeRosier, D.J. Three-dimensional structure of the frozen-hydrated flagellar filament. J. Mol. Biol. 1987, 195, 581–601.
- Wang, F.; Burrage, A.M.; Postel, S.; Clark, R.E.; Orlova, A.; Sundberg, E.J.; Kearns, D.B.; Egelman, E.H. A structural model of flagellar filament switching across multiple bacterial species. Nat. Commun. 2017, 8, 960.
- Mimori, Y.; Yamashita, I.; Murata, K.; Fujiyoshi, Y.; Yonekura, K.; Toyoshima, C.; Namba, K. The structure of the R-type straight flagellar filament of Salmonella at 9 A resolution by electron cryomicroscopy. J. Mol. Biol. 1995, 249, 69–87.
- Morgan, D.G.; Owen, C.; Melanson, L.A.; DeRosier, D.J. Structure of bacterial flagellar filaments at 11 A resolution: Packing of the alpha-helices. J. Mol. Biol. 1995, 249, 88–110.
- Mimori-Kiyosue, Y.; Yamashita, I.; Fujiyoshi, Y.; Yamaguchi, S.; Namba, K. Role of the outermost subdomain of Salmonella flagellin in the filament structure revealed by electron cryomicroscopy. J. Mol. Biol. 1998, 284, 521–530.
- Maki-Yonekura, S.; Yonekura, K.; Namba, K. Conformational change of flagellin for polymorphic supercoiling of the flagellar filament. Nat. Struct. Mol. Biol. 2010, 17, 417–422.
- Yamashita, I.; Hasegawa, K.; Suzuki, H.; Vonderviszt, F.; Mimori-Kiyosue, Y.; Namba, K. Structure and switching of bacterial flagellar filaments studied by X-ray fiber diffraction. Nat. Struct. Biol. 1998, 5, 125–132.
- Kitao, A.; Yonekura, K.; Maki-Yonekura, S.; Samatey, F.A.; Imada, K.; Namba, K.; Go, N. Switch interactions control energy frustration and multiple flagellar filament structures. Proc. Natl. Acad. Sci. USA 2006, 103, 4894–4899.
- Vonderviszt, F.; Imada, K.; Furukawa, Y.; Uedaira, H.; Taniguchi, H.; Namba, K. Mechanism of self-association and filament capping by flagellar HAP2. J. Mol. Biol. 1998, 284, 1399–1416.
- Postel, S.; Deredge, D.; Bonsor, D.A.; Yu, X.; Diederichs, K.; Helmsing, S.; Vromen, A.; Friedler, A.; Hust, M.; Egelman, E.H.; et al. Bacterial flagellar capping proteins adopt diverse oligomeric states. Elife 2016, 5, e18857.
- Song, W.S.; Cho, S.Y.; Hong, H.J.; Park, S.C.; Yoon, S.I. Self-Oligomerizing Structure of the Flagellar Cap Protein FliD and Its Implication in Filament Assembly. J. Mol. Biol. 2017, 429, 847–857.
- Cho, S.Y.; Song, W.S.; Hong, H.J.; Lee, G.S.; Kang, S.G.; Ko, H.J.; Kim, P.H.; Yoon, S.I. Tetrameric structure of the flagellar cap protein FliD from Serratia marcescens. Biochem. Biophys. Res. Commun. 2017, 489, 63–69.
- Cho, S.Y.; Song, W.S.; Oh, H.B.; Kim, H.U.; Jung, H.S.; Yoon, S.I. Structural analysis of the flagellar capping protein FliD from Helicobacter pylori. Biochem. Biophys. Res. Commun. 2019, 514, 98–104.
- Al-Otaibi, N.S.; Taylor, A.J.; Farrell, D.P.; Tzokov, S.B.; DiMaio, F.; Kelly, D.J.; Bergeron, J.R.C. The cryo-EM structure of the bacterial flagellum cap complex suggests a molecular mechanism for filament elongation. Nat. Commun. 2020, 11, 3210.
- Schuhmacher, J.S.; Thormann, K.M.; Bange, G. How bacteria maintain location and number of flagella? FEMS Microbiol. Rev. 2015, 39, 812–822.
- Chevance, F.F.; Hughes, K.T. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 2008, 6, 455–465.
- Wilhelms, M.; Molero, R.; Shaw, J.G.; Tomas, J.M.; Merino, S. Transcriptional hierarchy of Aeromonas hydrophila polar-flagellum genes. J. Bacteriol. 2011, 193, 5179–5190.
- Dasgupta, N.; Wolfgang, M.C.; Goodman, A.L.; Arora, S.K.; Jyot, J.; Lory, S.; Ramphal, R. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 2003, 50, 809–824.
- McCarter, L.L. Regulation of flagella. Curr. Opin. Microbiol. 2006, 9, 180–186.
- Jang, M.S.; Mouri, Y.; Uchida, K.; Aizawa, S.; Hayakawa, M.; Fujita, N.; Tezuka, T.; Ohnishi, Y. Genetic and Transcriptional Analyses of the Flagellar Gene Cluster in Actinoplanes missouriensis. J. Bacteriol. 2016, 198, 2219–2227.
- Ikeda, T.; Oosawa, K.; Hotani, H. Self-assembly of the filament capping protein, FliD, of bacterial flagella into an annular structure. J. Mol. Biol. 1996, 259, 679–686.
- Aldridge, P.; Gnerer, J.; Karlinsey, J.E.; Hughes, K.T. Transcriptional and translational control of the Salmonella fliC gene. J. Bacteriol. 2006, 188, 4487–4496.
- Koirala, S.; Mears, P.; Sim, M.; Golding, I.; Chemla, Y.R.; Aldridge, P.D.; Rao, C.V. A nutrient-tunable bistable switch controls motility in Salmonella enterica serovar Typhimurium. MBio 2014, 5, e01611–e01614.
- Wang, X.; Koirala, S.; Aldridge, P.D.; Rao, C.V. Two Tandem Mechanisms Control Bimodal Expression of the Flagellar Genes in Salmonella enterica. J. Bacteriol. 2020, 202, e00787-19.
- Niehus, E.; Gressmann, H.; Ye, F.; Schlapbach, R.; Dehio, M.; Dehio, C.; Stack, A.; Meyer, T.F.; Suerbaum, S.; Josenhans, C. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 2004, 52, 947–961.
- Alm, R.A.; Guerry, P.; Trust, T.J. Significance of duplicated flagellin genes in Campylobacter. J. Mol. Biol. 1993, 230, 359–363.
- Prouty, M.G.; Correa, N.E.; Klose, K.E. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 2001, 39, 1595–1609.
- Neville, B.A.; Sheridan, P.O.; Harris, H.M.; Coughlan, S.; Flint, H.J.; Duncan, S.H.; Jeffery, I.B.; Claesson, M.J.; Ross, R.P.; Scott, K.P.; et al. Pro-inflammatory flagellin proteins of prevalent motile commensal bacteria are variably abundant in the intestinal microbiome of elderly humans. PLoS ONE 2013, 8, e68919.
- Chen, Y.; Chai, Y.; Guo, J.H.; Losick, R. Evidence for cyclic Di-GMP-mediated signaling in Bacillus subtilis. J. Bacteriol. 2012, 194, 5080–5090.
- Purcell, E.B.; McKee, R.W.; McBride, S.M.; Waters, C.M.; Tamayo, R. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol. 2012, 194, 3307–3316.
- Simm, R.; Morr, M.; Kader, A.; Nimtz, M.; Romling, U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 2004, 53, 1123–1134.
- Kuchma, S.L.; Brothers, K.M.; Merritt, J.H.; Liberati, N.T.; Ausubel, F.M.; O’Toole, G.A. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 2007, 189, 8165–8178.
- Paul, K.; Nieto, V.; Carlquist, W.C.; Blair, D.F.; Harshey, R.M. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 2010, 38, 128–139.
- Fu, Y.; Yu, Z.; Liu, S.; Chen, B.; Zhu, L.; Li, Z.; Chou, S.H.; He, J. c-di-GMP Regulates Various Phenotypes and Insecticidal Activity of Gram-Positive Bacillus thuringiensis. Front. Microbiol. 2018, 9, 45.
- Verberkmoes, N.C.; Russell, A.L.; Shah, M.; Godzik, A.; Rosenquist, M.; Halfvarson, J.; Lefsrud, M.G.; Apajalahti, J.; Tysk, C.; Hettich, R.L.; et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009, 3, 179–189.
- Cullender, T.C.; Chassaing, B.; Janzon, A.; Kumar, K.; Muller, C.E.; Werner, J.J.; Angenent, L.T.; Bell, M.E.; Hay, A.G.; Peterson, D.A.; et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 2013, 14, 571–581.
- Gauger, E.J.; Leatham, M.P.; Mercado-Lubo, R.; Laux, D.C.; Conway, T.; Cohen, P.S. Role of motility and the flhDC Operon in Escherichia coli MG1655 colonization of the mouse intestine. Infect. Immun. 2007, 75, 3315–3324.
- Eriksson, S.; Lucchini, S.; Thompson, A.; Rhen, M.; Hinton, J.C. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 2003, 47, 103–118.
- Ott, M.; Messner, P.; Heesemann, J.; Marre, R.; Hacker, J. Temperature-dependent expression of flagella in Legionella. J. Gen. Microbiol. 1991, 137, 1955–1961.
- Kamp, H.D.; Higgins, D.E. A protein thermometer controls temperature-dependent transcription of flagellar motility genes in Listeria monocytogenes. PLoS Pathog. 2011, 7, e1002153.
- Anderson, P.E.; Gober, J.W. FlbT, the post-transcriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5′ untranslated region of flagellin mRNA. Mol. Microbiol. 2000, 38, 41–52.
- Yamamoto, S.; Kutsukake, K. FljA-mediated posttranscriptional control of phase 1 flagellin expression in flagellar phase variation of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2006, 188, 958–967.
- Yakhnin, H.; Pandit, P.; Petty, T.J.; Baker, C.S.; Romeo, T.; Babitzke, P. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol. Microbiol. 2007, 64, 1605–1620.
- Anderson, D.K.; Newton, A. Posttranscriptional regulation of Caulobacter flagellin genes by a late flagellum assembly checkpoint. J. Bacteriol. 1997, 179, 2281–2288.
- Sal, M.S.; Li, C.; Motalab, M.A.; Shibata, S.; Aizawa, S.; Charon, N.W. Borrelia burgdorferi uniquely regulates its motility genes and has an intricate flagellar hook-basal body structure. J. Bacteriol. 2008, 190, 1912–1921.
- Mukherjee, S.; Yakhnin, H.; Kysela, D.; Sokoloski, J.; Babitzke, P.; Kearns, D.B. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol. Microbiol. 2011, 82, 447–461.
- Khanra, N.; Rossi, P.; Economou, A.; Kalodimos, C.G. Recognition and targeting mechanisms by chaperones in flagellum assembly and operation. Proc. Natl. Acad. Sci. USA 2016, 113, 9798–9803.
- Auvray, F.; Thomas, J.; Fraser, G.M.; Hughes, C. Flagellin polymerisation control by a cytosolic export chaperone. J. Mol. Biol. 2001, 308, 221–229.
- Ozin, A.J.; Claret, L.; Auvray, F.; Hughes, C. The FliS chaperone selectively binds the disordered flagellin C-terminal D0 domain central to polymerisation. FEMS Microbiol. Lett. 2003, 219, 219–224.
- Muskotal, A.; Kiraly, R.; Sebestyen, A.; Gugolya, Z.; Vegh, B.M.; Vonderviszt, F. Interaction of FliS flagellar chaperone with flagellin. FEBS Lett. 2006, 580, 3916–3920.
- Evdokimov, A.G.; Phan, J.; Tropea, J.E.; Routzahn, K.M.; Peters, H.K.; Pokross, M.; Waugh, D.S. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat. Struct. Biol. 2003, 10, 789–793.
- Galeva, A.; Moroz, N.; Yoon, Y.H.; Hughes, K.T.; Samatey, F.A.; Kostyukova, A.S. Bacterial flagellin-specific chaperone FliS interacts with anti-sigma factor FlgM. J. Bacteriol. 2014, 196, 1215–1221.
- Furukawa, Y.; Inoue, Y.; Sakaguchi, A.; Mori, Y.; Fukumura, T.; Miyata, T.; Namba, K.; Minamino, T. Structural stability of flagellin subunit affects the rate of flagellin export in the absence of FliS chaperone. Mol. Microbiol. 2016, 102, 405–416.
- Bange, G.; Kummerer, N.; Engel, C.; Bozkurt, G.; Wild, K.; Sinning, I. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc. Natl. Acad. Sci. USA 2010, 107, 11295–11300.
- Kinoshita, M.; Hara, N.; Imada, K.; Namba, K.; Minamino, T. Interactions of bacterial flagellar chaperone-substrate complexes with FlhA contribute to co-ordinating assembly of the flagellar filament. Mol. Microbiol. 2013, 90, 1249–1261.
- Terahara, N.; Inoue, Y.; Kodera, N.; Morimoto, Y.V.; Uchihashi, T.; Imada, K.; Ando, T.; Namba, K.; Minamino, T. Insight into structural remodeling of the FlhA ring responsible for bacterial flagellar type III protein export. Sci. Adv. 2018, 4, eaao7054.
- Xing, Q.; Shi, K.; Portaliou, A.; Rossi, P.; Economou, A.; Kalodimos, C.G. Structures of chaperone-substrate complexes docked onto the export gate in a type III secretion system. Nat. Commun. 2018, 9, 1773.
- Altegoer, F.; Mukherjee, S.; Steinchen, W.; Bedrunka, P.; Linne, U.; Kearns, D.B.; Bange, G. FliS/flagellin/FliW heterotrimer couples type III secretion and flagellin homeostasis. Sci. Rep. 2018, 8, 11552.
- Yokoseki, T.; Iino, T.; Kutsukake, K. Negative regulation by fliD, fliS, and fliT of the export of the flagellum-specific anti-sigma factor, FlgM, in Salmonella typhimurium. J. Bacteriol. 1996, 178, 899–901.
- Yokoseki, T.; Kutsukake, K.; Ohnishi, K.; Iino, T. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology (Reading) 1995, 141, 1715–1722.
- Xu, S.; Peng, Z.; Cui, B.; Wang, T.; Song, Y.; Zhang, L.; Wei, G.; Wang, Y.; Shen, X. FliS modulates FlgM activity by acting as a non-canonical chaperone to control late flagellar gene expression, motility and biofilm formation in Yersinia pseudotuberculosis. Environ. Microbiol. 2014, 16, 1090–1104.
- Imada, K.; Minamino, T.; Kinoshita, M.; Furukawa, Y.; Namba, K. Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners. Proc. Natl. Acad. Sci. USA 2010, 107, 8812–8817.




