
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Diana Martonik | + 2404 word(s) | 2404 | 2021-06-21 04:23:11 | | | |
| 2 | Bruce Ren | -21 word(s) | 2383 | 2021-06-25 08:24:01 | | |
Video Upload Options
COVID-19 is an acute infectious disease of the respiratory system caused by infection with the SARS-CoV-2 virus (Severe Acute Respiratory Syndrome Coronavirus 2). Transmission of SARS-CoV-2 infections occurs through droplets and contaminated objects. A rapid and well-coordinated immune system response is the first line of defense in a viral infection. However, a disturbed and over-activated immune response may be counterproductive, causing damage to the body. Severely ill patients hospitalised with COVID-19 exhibit increased levels of many cytokines, including Interleukin (IL)-1β, IL-2, IL-6, IL-7, IL-8, IL-10, IL-17, granulocyte colony stimulating factor (G-CSF), monocyte chemoattractant protein 1 (MCP-1) and tumor necrosis factor (TNF). Increasing evidence suggests that Th17 cells play an important role in the pathogenesis of COVID-19, not only by activating cytokine cascade but also by inducing Th2 responses, inhibiting Th1 differentiation and suppressing Treg cells.
1. Introduction
2. Mechanisms of SARS-CoV-2 Invasion
3. T Cells Differentiation
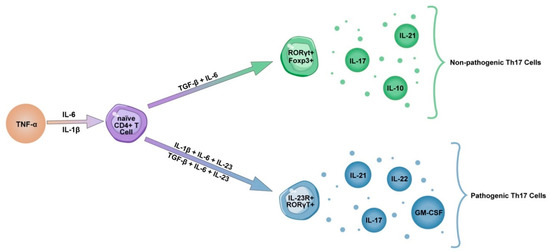
4. Th17 Lineage
5. IL-23 Influence on Th17 Pathogenicity
6. Th17 and Tc17 Cells in COVID-19
References
- Catanzaro, M.; Fagiani, F.; Racchi, M.; Corsini, E.; Govoni, S.; Lanni, C. Immune response in COVID-19: Addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct. Target. Ther. 2020, 5, 1–10.
- Chan, J.F.W.; Yuan, S.; Kok, K.H.; To, K.K.W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.Y.; Poon, R.W.S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523.
- Tang, Y.; Liu, J.; Zhang, D.; Xu, Z.; Ji, J.; Wen, C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020, 11, 1708.
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.
- Akçay, Ş.; Özlü, T.; Yilmaz, A. Radiological approaches to COVID-19 pneumonia. Turk. J. Med. Sci. 2020, 50, 604–610.
- Hu, L.; Wang, C. Radiological role in the detection, diagnosis and monitoring for the coronavirus disease 2019 (COVID-19). Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4523–4528.
- Hojyo, S.; Uchida, M.; Tanaka, K.; Hasebe, R.; Tanaka, Y.; Murakami, M.; Hirano, T. How COVID-19 induces cytokine storm with high mortality. Inflamm. Regen. 2020, 40, 1–7.
- Bordoni, V.; Sacchi, A.; Cimini, E.; Notari, S.; Grassi, G.; Tartaglia, E.; Casetti, R.; Giancola, M.L.; Bevilacqua, N.; Maeurer, M.; et al. An Inflammatory Profile Correlates with Decreased Frequency of Cytotoxic Cells in Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2272–2275.
- Li, H.; Liu, L.; Zhang, D.; Xu, J.; Dai, H.; Tang, N.; Su, X.; Cao, B. SARS-CoV-2 and viral sepsis: Observations and hypotheses. Lancet 2020, 395, 1517–1520.
- Tay, M.Z.; Poh, C.M.; Rénia, L.; Macary, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374.
- De Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; Tartaro, D.L.; Mattioli, M.; et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020, 11, 1–17.
- Schultheiß, C.; Paschold, L.; Simnica, D.; Mohme, M.; Willscher, E.; von Wenserski, L.; Scholz, R.; Wieters, I.; Dahlke, C.; Tolosa, E.; et al. Next-Generation Sequencing of T and B Cell Receptor Repertoires from COVID-19 Patients Showed Signatures Associated with Severity of Disease. Immunity 2020, 53, 442–455.
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613.
- Toh, M.-L.; Kawashima, M.; Zrioual, S.; Hot, A.; Miossec, P.; Miossec, P. IL-17 inhibits human Th1 differentiation through IL-12Rβ2 downregulation. Cytokine 2009, 48, 226–230.
- Wang, Y.-H.; Liu, Y.-J. The IL-17 cytokine family and their role in allergic inflammation. Curr. Opin. Immunol. 2008, 20, 697–702.
- Costa, L.B.; Perez, L.G.; Palmeira, V.A.; E Cordeiro, T.M.; Ribeiro, V.T.; Lanza, K.; E Silva, A.C.S. Insights on SARS-CoV-2 Molecular Interactions with the Renin-Angiotensin System. Front. Cell Dev. Biol. 2020, 8, 559841.
- Kreutzberger, A.J.B.; Sanyal, A.; Ojha, R.; Pyle, J.D.; Vapalahti, O.; Balistreri, G.; Kirchhausen, T. Synergistic inhibition of two host factors that facilitate entry of Severe Acute Respiratory Syndrome Coronavirus 2. bioRxiv 2021.
- Chu, H.; Chan, J.F.-W.; Yuen, T.T.-T.; Shuai, H.; Yuan, S.; Wang, Y.; Hu, B.; Yip, C.C.-Y.; Tsang, J.O.-L.; Huang, X.; et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: An observational study. Lancet Microbe 2020, 1, e14–e23.
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263.
- Anguiano, L.; Riera, M.; Pascual, J.; Soler, M.J. Circulating ACE2 in Cardiovascular and Kidney Diseases. Curr. Med. Chem. 2017, 24, 3231–3241.
- Touyz, R.M.; Montezano, A.C. Angiotensin-(1–7) and Vascular Function. Hypertension 2018, 71, 68–69.
- Benigni, A.; Cassis, P.; Remuzzi, G. Angiotensin II revisited: New roles in inflammation, immunology and aging. EMBO Mol. Med. 2010, 2, 247–257.
- Chung, M.K.; Karnik, S.; Saef, J.; Bergmann, C.; Barnard, J.; Lederman, M.M.; Tilton, J.; Cheng, F.; Harding, C.V.; Young, J.B.; et al. SARS-CoV-2 and ACE2: The biology and clinical data settling the ARB and ACEI controversy. EBioMedicine 2020, 58, 102907.
- Murphy, K.M.; Ouyang, W.; Farrar, J.D.; Yang, J.; Ranganath, S.; Asnagli, H.; Afkarian, M.; Murphy, T.L. Signaling and Transcription in T Helper Development. Annu. Rev. Immunol. 2000, 18, 451–494.
- Luckheeram, R.V.; Zhou, R.; Verma, A.D.; Xia, B. CD4+T Cells: Differentiation and Functions. Clin. Dev. Immunol. 2012, 2012, 925135.
- Dardalhon, V.; Korn, T.; Kuchroo, V.K.; Anderson, A.C. Role of Th1 and Th17 cells in organ-specific autoimmunity. J. Autoimmun. 2008, 31, 252–256.
- Paul, W.E.; Zhu, J. How are TH2-type immune responses initiated and amplified? Nat. Rev. Immunol. 2010, 10, 225–235.
- Neidleman, J.; Luo, X.; Frouard, J.; Xie, G.; Gill, G.; Stein, E.S.; McGregor, M.; Ma, T.; George, A.F.; Kosters, A.; et al. SARS-CoV-2-Specific T Cells Exhibit Phenotypic Features of Helper Function, Lack of Terminal Differentiation, and High Proliferation Potential. Cell Rep. Med. 2020, 1, 6, 100081.
- Hotez, P.J.; Bottazzi, M.E.; Corry, D.B. The potential role of Th17 immune responses in coronavirus immunopathology and vaccine-induced immune enhancement. Microbes Infect. 2020, 22, 165–167.
- Lee, G.R. The balance of Th17 versus treg cells in autoimmunity. Int. J. Mol. Sci. 2018, 19, 730.
- Ali, N.; Rosenblum, M.D. Regulatory T cells in skin. Immunology 2017, 152, 372–381.
- Bedoya, S.K.; Lam, B.; Lau, K.; Larkin, J. Th17 Cells in Immunity and Autoimmunity. Clin. Dev. Immunol. 2013, 2013, 986789.
- Gaffen, S.L.; Jain, R.; Garg, A.V.; Cua, D.J. The IL-23–IL-17 immune axis: From mechanisms to therapeutic testing. Nat. Rev. Immunol. 2014, 14, 585–600.
- Gu, C.; Wu, L.; Li, X. IL-17 family: Cytokines, receptors and signaling. Cytokine 2013, 64, 477–485.
- Muyayalo, K.P.; Huang, D.; Zhao, S.; Xie, T.; Mor, G.; Liao, A. COVID-19 and Treg/Th17 imbalance: Potential relationship to pregnancy outcomes. Am. J. Reprod. Immunol. 2020, 84, e13304.
- McGeachy, M.J.; Cua, D.J.; Gaffen, S.L. The IL-17 Family of Cytokines in Health and Disease. Immunity 2019, 50, 892–906.
- Liu, S.M.; King, C. IL-21–Producing Th Cells in Immunity and Autoimmunity. J. Immunol. 2013, 191, 3501–3506.
- Spolski, R.; Leonard, W.J. Interleukin-21: A double-edged sword with therapeutic potential. Nat. Rev. Drug Discov. 2014, 13, 379–395.
- Long, D.; Chen, Y.; Wu, H.; Zhao, M.; Lu, Q. Clinical significance and immunobiology of IL-21 in autoimmunity. J. Autoimmun. 2019, 99, 1–14.
- Tian, Y.; Zajac, A.J. IL-21 and T Cell Differentiation: Consider the Context. Trends Immunol. 2016, 37, 557–568.
- Dudakov, J.A.; Hanash, A.M.; Brink, M.R.V.D. Interleukin-22: Immunobiology and Pathology. Annu. Rev. Immunol. 2015, 33, 747–785.
- Parks, O.B.; Pociask, D.A.; Hodzic, Z.; Kolls, J.K.; Good, M. Interleukin-22 Signaling in the Regulation of Intestinal Health and Disease. Front. Cell Dev. Biol. 2016, 3, 85.
- Eyerich, K.; DiMartino, V.; Cavani, A. IL-17 and IL-22 in immunity: Driving protection and pathology. Eur. J. Immunol. 2017, 47, 607–614.
- Shiomi, A.; Usui, T.; Mimori, T. GM-CSF as a therapeutic target in autoimmune diseases. Inflamm. Regen. 2016, 36, 1–9.
- Becher, B.; Tugues, S.; Greter, M. GM-CSF: From Growth Factor to Central Mediator of Tissue Inflammation. Immunity 2016, 45, 963–973.
- Aliper, A.M.; Frieden-Korovkina, V.P.; Buzdin, A.; Roumiantsev, S.; Zhavoronkov, A. A role for G-CSF and GM-CSF in nonmyeloid cancers. Cancer Med. 2014, 3, 737–746.
- Avci, A.B.; Feist, E.; Burmester, G.-R. Targeting GM-CSF in rheumatoid arthritis. Clin. Exp. Rheumatol. 2016, 34, 39–44.
- Tau, G.; Rothman, P.B. Biologic functions of the IFN-gamma receptors. Allergy 1999, 54, 1233–1251.
- Ivashkiv, L.B. IFNγ: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 545–558.
- Kak, G.; Raza, M.; Tiwari, B.K. Interferon-gamma (IFN-γ): Exploring its implications in infectious diseases. Biomol. Concepts 2018, 9, 64–79.
- Ishikawa, Y.; Iwata, S.; Hanami, K.; Nawata, A.; Zhang, M.; Yamagata, K.; Hirata, S.; Sakata, K.; Todoroki, Y.; Nakano, K.; et al. Relevance of interferon-gamma in pathogenesis of life-threatening rapidly progressive interstitial lung disease in patients with dermatomyositis. Arthritis Res. 2018, 20, 1–10.
- Bettelli, E.; Korn, T.; Oukka, M.; Kuchroo, V.K. Induction and effector functions of TH17 cells. Nat. Cell Biol. 2008, 453, 1051–1057.
- McGeachy, M.J.; Bak-Jensen, K.S.; Chen, Y.; Tato, C.M.; Blumenschein, W.M.; McClanahan, T.K.; Cua, D.J. TGF-β and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain TH-17 cell–mediated pathology. Nat. Immunol. 2007, 8, 1390–1397.
- Lee, Y.; Awasthi, A.; Yosef, N.; Quintana, F.J.; Xiao, S.; Peters, A.; Wu, C.; Kleinewietfeld, M.; Kunder, S.; Hafler, D.A.; et al. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 2012, 13, 991–999.
- Bettelli, E.; Korn, T.; Kuchroo, V.K. Th17: The third member of the effector T cell trilogy. Curr. Opin. Immunol. 2007, 19, 652–657.
- Tsukazaki, H.; Kaito, T. The Role of the IL-23/IL-17 Pathway in the Pathogenesis of Spondyloarthritis. Int. J. Mol. Sci. 2020, 21, 6401.
- Pastor-Fernández, G.; Mariblanca, I.R.; Navarro, M.N. Decoding IL-23 Signaling Cascade for New Therapeutic Opportunities. Cells 2020, 9, 2044.
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768.
- Neurath, M.F. COVID-19 and immunomodulation in IBD. Gut 2020, 69, 1335–1342.
- Lin, S.; Wu, H.; Wang, C.; Xiao, Z.; Xu, F. Regulatory T Cells and Acute Lung Injury: Cytokines, Uncontrolled Inflammation, and Therapeutic Implications. Front. Immunol. 2018, 9, 1545.
- Yu, Z.-X.; Ji, M.-S.; Yan, J.; Cai, Y.; Liu, J.; Yang, H.-F.; Li, Y.; Jin, Z.-C.; Zheng, J.-X. The ratio of Th17/Treg cells as a risk indicator in early acute respiratory distress syndrome. Crit. Care 2015, 19, 82.
- Wu, D.; Yang, X.O. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020, 53, 368–370.
- Burgos-Blasco, B.; Güemes-Villahoz, N.; Santiago, J.L.; Fernandez-Vigo, J.I.; Espino-Paisán, L.; Sarriá, B.; García-Feijoo, J.; Martinez-De-La-Casa, J.M. Hypercytokinemia in COVID-19: Tear cytokine profile in hospitalized COVID-19 patients. Exp. Eye Res. 2020, 200, 108253.
- Petrone, L.; Petruccioli, E.; Vanini, V.; Cuzzi, G.; Fard, S.N.; Alonzi, T.; Castilletti, C.; Palmieri, F.; Gualano, G.; Vittozzi, P.; et al. A whole blood test to measure SARS-CoV-2-specific response in COVID-19 patients. Clin. Microbiol. Infect. 2021, 27, 286.e7–286.e13.
- Ghazavi, A.; Ganji, A.; Keshavarzian, N.; Rabiemajd, S.; Mosayebi, G. Cytokine profile and disease severity in patients with COVID-19. Cytokine 2021, 137, 155323.
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.A.; Endeman, H.; Akker, J.P.C.V.D.; Molenkamp, R.; Koopmans, M.P.G.; Van Gorp, E.C.M.; et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5, eabd2071.
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422.
- Pasrija, R.; Naime, M. The deregulated immune reaction and cytokines release storm (CRS) in COVID-19 disease. Int. Immunopharmacol. 2021, 90, 107225.
- Ronit, A.; Berg, R.M.; Bay, J.T.; Haugaard, A.K.; Ahlström, M.G.; Burgdorf, K.S.; Ullum, H.; Rørvig, S.B.; Tjelle, K.; Foss, N.B.; et al. Compartmental immunophenotyping in COVID-19 ARDS: A case series. J. Allergy Clin. Immunol. 2021, 147, 81–91.
- Mahallawi, W.H.; Khabour, O.F.; Zhang, Q.; Makhdoum, H.M.; Suliman, B.A. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine 2018, 104, 8–13.
- Faure, E.; Poissy, J.; Goffard, A.; Fournier, C.; Kipnis, E.; Titecat, M.; Bortolotti, P.; Martinez, L.; Dubucquoi, S.; Dessein, R.; et al. Distinct Immune Response in Two MERS-CoV-Infected Patients: Can We Go from Bench to Bedside? PLoS ONE 2014, 9, e88716.
- Josset, L.; Menachery, V.D.; Gralinski, L.E.; Agnihothram, S.; Sova, P.; Carter, V.S.; Yount, B.L.; Graham, R.L.; Baric, R.S.; Katze, M.G. Cell Host Response to Infection with Novel Human Coronavirus EMC Predicts Potential Antivirals and Important Differences with SARS Coronavirus. mBio 2013, 4, e00165-13.
- Bermejo-Martin, J.F.; De Lejarazu, R.O.; Pumarola, T.; Rello, J.; Almansa, R.; Ramírez, P.; Martin-Loeches, I.; Varillas, D.; Gallegos, M.C.; Serón, C.; et al. Th1 and Th17 hypercytokinemia as early host response signature in severe pandemic influenza. Crit. Care 2009, 13, R201.
- Mikacenic, C.; Hansen, E.E.; Radella, F.; Gharib, S.A.; Stapleton, R.D.; Wurfel, M.M. Interleukin-17A Is Associated With Alveolar Inflammation and Poor Outcomes in Acute Respiratory Distress Syndrome. Crit. Care Med. 2016, 44, 496–502.
- Zheng, L.-Y.; Sun, P.-C. Increased Expression of IL-23 and IL-17 in Serum of Patients with Neonatal Respiratory Distress Syndrome and its Clinical Significance. Clin. Lab. 2020, 66, 8.
- Ding, Q.; Liu, G.-Q.; Zeng, Y.-Y.; Zhu, J.-J.; Liu, Z.-Y.; Zhang, X.; Huang, J.-A. Role of IL-17 in LPS-induced acute lung injury: An in vivo study. Oncotarget 2017, 8, 93704–93711.
- Rout, N. Enhanced Th1/Th17 Functions of CD161+ CD8+ T Cells in Mucosal Tissues of Rhesus Macaques. PLoS ONE 2016, 11, e0157407.
- McCarthy, M.K.; Zhu, L.; Procario, M.C.; Weinberg, J.B. IL-17 contributes to neutrophil recruitment but not to control of viral replication during acute mouse adenovirus type 1 respiratory infection. Virology 2014, 456–457, 259–267.
- Lindén, A.; Laan, M.; Anderson, G.P. Neutrophils, interleukin-17A and lung disease. Eur. Respir. J. 2005, 25, 159–172.
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine 2020, 55, 102763.
- Han, H.; Ma, Q.; Li, C.; Liu, R.; Zhao, L.; Wang, W.; Zhang, P.; Liu, X.; Gao, G.; Liu, F.; et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 2020, 9, 1123–1130.
- Yao, Z.; Zheng, Z.; Wu, K.; Junhua, Z. Immune environment modulation in pneumonia patients caused by coronavirus: SARS-CoV, MERS-CoV and SARS-CoV-2. Aging 2020, 12, 7639–7651.
- Croci, S.; Bonacini, M.; Dolci, G.; Massari, M.; Facciolongo, N.; Pignatti, E.; Pisciotta, A.; Carnevale, G.; Negro, A.; Cassone, G.; et al. Human Dental Pulp Stem Cells Modulate Cytokine Production in vitro by Peripheral Blood Mononuclear Cells from Coronavirus Disease 2019 Patients. Front. Cell Dev. Biol. 2021, 8, 609204.
- Lucas, C.; Team, Y.I.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nat. Cell Biol. 2020, 584, 463–469.
- Sakaguchi, R.; Chikuma, S.; Shichita, T.; Morita, R.; Sekiya, T.; Ouyang, W.; Ueda, T.; Seki, H.; Morisaki, H.; Yoshimura, A. Innate-like function of memory Th17 cells for enhancing endotoxin-induced acute lung inflammation through IL-22. Int. Immunol. 2015, 28, 233–243.
- Bonny, T.S.; Patel, E.U.; Zhu, X.; Bloch, E.M.; Grabowski, M.K.; Abraham, A.G.; Littlefield, K.; Shrestha, R.; E Benner, S.; Laeyendecker, O.; et al. Cytokine and Chemokine Levels in Coronavirus Disease 2019 Convalescent Plasma. Open Forum Infect. Dis. 2021, 8, ofaa574.




