
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Baltasar Mayo | + 4073 word(s) | 4073 | 2020-04-23 10:24:41 | | | |
| 2 | Rita Xu | -867 word(s) | 3206 | 2020-04-26 09:27:15 | | | | |
| 3 | Rita Xu | -96 word(s) | 3110 | 2020-10-29 09:10:09 | | |
Video Upload Options
During the selection process of probiotics for vaginal applications, twenty-five lactic acid bacteria (LAB) isolates from human vagina belonging to six different species were tested for antimicrobial resistance by a microdilution method. Gene-specific PCR amplifications proved the strains carry no acquired antibiotic resistance genes, except for a tet(W) gene present in two tetracycline-susceptible Bifidobacterium bifidum strains. Genome analysis of a selected set of strains showed no other acquired resistance determinants. The tet(W) of B. bifidum was inactive by the insertion of two guanine residues in the middle of the gene. Surprisingly, the inactive gene became active and functional very easily, providing resistance to tetracycline and remaining stable afterward. LAB intended to be used in health applications must be free of acquired antimicrobial resistance genes; these could be spread and transferred to human pathogens.
1. Isolation, Identification and Typing of Vaginal LAB
Twenty-five vaginal LAB isolates with clear acidification halos on MRS agar supplemented with 0.5% CaCO3 were recovered. All were Gram-positive rods, catalase negative, and γ-hemolytic. They were subsequently identified at the species level by 16S rRNA gene sequencing and sequence comparison. Seven isolates were identified as belonging to Lactobacillus crispatus, six to Lactobacillus salivarius, four to Lactobacillus jensenii, four to Lactobacillus paracasei, two to Lactobacillus reuteri, and two to Bifidobacterium bifidum (Table 1). A fingerprinting analysis by combining the results of RAPD-PCR and rep-PCR techniques detected 21 different strains among the 25 isolates, distributed as follows: L. crispatus (six strains), L. salivarius (five), L. jensenii (three), L. paracasei (three), L. reuteri (two), and B. bifidum (two strains) (Supplementary Figure S1).
Table 1. Minimum inhibitory concentration (MIC) values of 16 antibiotics to the vaginal LAB species and strains of this study.
| Species | Strain | Antibiotic (MIC as µg mL−1) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEN | KAN | STR | NEO | TET | ERY | CLI | CHL | AMP | PEN | VAN | QDA | LIN | TMP | CIP | RIF | ||
| L. crispatus | VA20-32AN | 2 | 16 | 2 | 16 | 2 | 0.06 | 0.25 | 4 | 1 | 0.5 | 0.5 | 1 | 4 | >64 | 16 | 1 |
| VA27-7 | 4 | 32 | 64 | 8 | 1 | 1 | 2 | 8 | 2 | 2 | 1 | 1 | 4 | 32 | 64 | 4 | |
| VA27-9 | 1 | 16 | 2 | 2 | 2 | 0.03 | 0.5 | 4 | 2 | 0.5 | 0.5 | 1 | 4 | 64 | 32 | 2 | |
| VA28-12 | 1 | 16 | 2 | 2 | 2 | 0.06 | 0.5 | 4 | 2 | 0.5 | 0.5 | 2 | 4 | 64 | 32 | 2 | |
| VA32-17 | 2 | 64 | 2 | 8 | 4 | 0.03 | 0.5 | 2 | 1 | 1 | 0.5 | 1 | 2 | >64 | 64 | 8 | |
| VA32-17AN | 4 | 128 | 32 | 4 | 2 | 0.25 | 0.5 | 4 | 1 | 0.5 | 1 | 1 | 2 | 16 | 32 | 4 | |
| VA50-4AN | ≤0.5 | 32 | 1 | 2 | 4 | 0.12 | 0.12 | 4 | 4 | 1 | 0.5 | 1 | 4 | >64 | 32 | 4 | |
| L. jensenii | VA04-1AN | ≤0.5 | 4 | 2 | 1 | 0.25 | ≤0.016 | 0.12 | 4 | 0.25 | 0.12 | 1 | 0.5 | 1 | >64 | 8 | 0.25 |
| VA04-2AN | ≤0.5 | 4 | 4 | 1 | 0.5 | 0.03 | 0.12 | 2 | 0.5 | 1 | 1 | 0.5 | 2 | >64 | 8 | 0.25 | |
| VA15-2AN | ≤0.5 | ≤2 | 1 | ≤0.5 | 1 | ≤0.016 | ≤0.03 | 2 | 0.06 | 0.06 | 0.5 | 0.5 | 0.5 | >64 | 8 | 0.25 | |
| VA16-11 | ≤0.5 | 8 | 1 | 2 | 4 | 0.06 | 0.25 | 4 | 0.06 | ≤0.03 | 2 | 0.5 | 2 | >64 | 16 | 0.5 | |
| Breakpoint (µg mL−1) | 16 | 16 | 16 | - | 4 | 1 | 4 | 4 | 2 | - | 2 | - | - | - | - | - | |
| L. salivarius | VA09-4 | 8 | 64 | 16 | 4 | 2 | 0.25 | 0.25 | 2 | 1 | 0.25 | 128 | 0.25 | 0.5 | ≤0.12 | 1 | 2 |
| VA16-20 | ≤0.5 | 4 | 2 | 0.5 | 1 | 0.06 | 0.06 | 2 | 0.5 | 0.12 | >128 | 0.5 | 0.5 | 0.25 | 0.5 | 0.5 | |
| VA37-13 | ≤0.5 | 4 | ≤0.5 | ≤0.5 | 0.5 | 0.06 | 0.06 | 2 | 0.25 | 0.12 | >128 | 0.5 | 0.5 | 0.25 | ≤0.25 | 0.5 | |
| VA40-10 | 128 | >1024 | >256 | 256 | 2 | 1 | 1 | 4 | 1 | 0.25 | >128 | 1 | 1 | 1 | 4 | 0.5 | |
| VA40-12AN | 4 | 128 | 32 | 4 | 2 | 0.25 | 0.25 | 4 | 0.5 | 0.25 | >128 | 1 | 0.5 | 0.25 | 1 | 1 | |
| VA40-14AN | 4 | 128 | 32 | 4 | 2 | 0.25 | 0.5 | 4 | 0.5 | 0.25 | >128 | 1 | 0.5 | ≤0.12 | 1 | 1 | |
| Breakpoint (µg mL−1) | 16 | 64 | 64 | - | 8 | 1 | 4 | 4 | 4 | - | n.r. | - | - | - | - | - | |
| L. paracasei | VA02-1AN | ≤0.5 | 16 | 8 | 1 | 2 | 0.12 | 0.06 | 8 | 1 | 0.25 | >128 | 1 | 4 | 0.5 | 4 | 0.5 |
| VA24-4 | 1 | 16 | 8 | 4 | 4 | 0.12 | 0.06 | 4 | 0.5 | 0.25 | >128 | 1 | 2 | 0.25 | 4 | 0.5 | |
| VA26-3 | ≤0.5 | 16 | 8 | 2 | 2 | 0.12 | 0.06 | 4 | 1 | 0.25 | >128 | 1 | 2 | 1 | 2 | 0.5 | |
| VA27-8 | 1 | 32 | 16 | 8 | 2 | 0.06 | 0.06 | 8 | 0.5 | 0.25 | >128 | 1 | 4 | 0.25 | 4 | 0.5 | |
| Breakpoint (µg mL−1) | 32 | 64 | 64 | - | 4 | 1 | 4 | 4 | 4 | - | n.r. | - | - | - | - | - | |
| L. reuteri | VA15-3 | ≤0.5 | 4 | 2 | ≤0.5 | 8 | 0.12 | ≤0.03 | 4 | 1 | 2 | >128 | 1 | 2 | >64 | 32 | 0.25 |
| VA24-5 | ≤0.5 | 16 | 4 | ≤0.5 | 16 | 0.06 | ≤0.03 | 4 | 2 | 8 | >128 | 0.5 | 4 | >64 | 32 | 0.25 | |
| Breakpoint (µg mL−1) | 8 | 64 | 64 | - | 32 | 1 | 4 | 4 | 2 | - | n.r. | - | - | - | - | - | |
| B. bifidum | VA07-1AN | 8 | 64 | >256 | 16 | 1 | ≤0.016 | 0.06 | 1 | ≤0.03 | ≤0.03 | 0.5 | 0.5 | 0.5 | 16 | 8 | 2 |
| VA07-2AN | 32 | 64 | >256 | 32 | 1 | ≤0.016 | ≤0.03 | 1 | ≤0.03 | ≤0.03 | 1 | 0.5 | 0.5 | 16 | 8 | 1 | |
| Breakpoint (µg mL−1) | 64 | - | 128 | - | 8 | 1 | 1 | 4 | 2 | - | 2 | - | - | - | - | - | |
2. Antibiotic Susceptibility
Table 1 shows the MIC values of the 16 tested antibiotics for the 25 vaginal LAB isolates. All isolates were phenotypically susceptible to tetracycline, erythromycin, clindamycin, penicillin, quinupristin-dalfopristin, linezolid, and rifampicin. The distribution of neomycin MICs covered more than nine 2-fold dilutions, ranging from ≤0.5 to 256 µg mL−1. Similarly, a wide distribution of MICs was observed for streptomycin (≤0.5 to >256 µg mL−1), ciprofloxacin (≤0.25 to 64 µg mL−1), and chloramphenicol (1 to 8 µg mL−1). The most common resistance phenotypes observed were those to trimethoprim (MIC 16 to ≥64 µg mL−1) and vancomycin (MIC ≥ 128 µg mL−1). Nine isolates were resistant to kanamycin (MIC values 32 to >1024 µg mL−1). The two B. bifidum strains were susceptible to all tested antibiotics except for streptomycin (MIC >256 µg mL−1). Moderate resistance to chloramphenicol was seen in three isolates and for ampicillin in one isolate. Interestingly, L. salivarius VA40-10 was highly resistant to all four aminoglycosides tested (gentamicin, kanamycin, streptomycin, and neomycin).
3. Detection of AR Genes by PCR
The presence of genes coding for the commonest AR genes spread among LAB was investigated by PCR. No genes involved in resistance to chloramphenicol (cat), β-lactams (bla), aminoglycosides [aac(6′)-aph(2″) and aad(E)], macrolides [erm(A), erm(B), erm(C), erm(F), mef(A)], tetracycline [tet(M), tet(O), tet(S), tet(K), tet(L)], clindamycin (lsaA) or vancomycin (vanA) were ever detected in any of the isolates (data now shown). In contrast, PCR analyses for genes encoding ribosomal protection proteins (RPP) causing tetracycline resistance using the degenerate primer pairs DI-DII and Tet1-Tet2 (Supplementary Table S1) produced a positive amplification when DNA from two tetracycline-susceptible B. bifidum isolates was used as a template (Figure 1A). Amplification with gene-specific primers gave a positive result only for the tet(W) gene (Figure 1B). Amplicon sequencing and sequence comparison further proved the presence in these isolates of a tet(W) gene highly homologous to those present in many Gram-positive and Gram-negative bacteria.
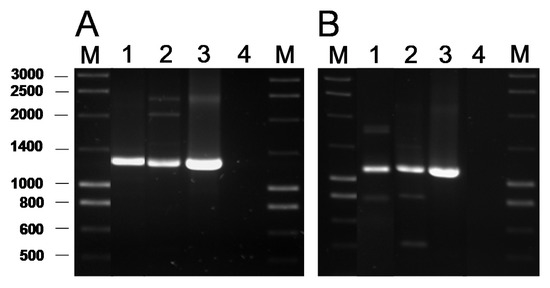
Figure 1. PCR amplification of tetracycline resistance genes using the universal primers Tet1 and Tet2 targeting a segment of 1,300 bp of the genes encoding RPP (A) and 1,200 bp of the tet(W) gene with the specific primer pair tetWF-Tet2 (B). Key of samples: Lane 1, DNA from B. bifidum VA07-1AN; lane 2, B. bifidum VA07-2AN; lane 3, Leuconostoc mesenteroides subsp. mesenteroides LbE16 (positive control) [1]; line 4, blank (no template DNA). M, molecular weight marker.
4. Genome Analysis for AR Genes
Based on the phenotype and genotype results (Table 1; Supplementary Figure S1), six strains were subjected to genome sequencing: L. crispatus VA50-4AN (resistant to kanamycin, ampicillin, and trimethoprim), L. jensenii VA04-2AN (resistant to trimethoprim), L. salivarius VA40-10 (resistant to gentamicin, kanamycin, streptomycin, neomycin, and vancomycin), L. paracasei VA02-1AN (resistant to chloramphenicol and vancomycin), L. reuteri VA24-5 (resistant to vancomycin and trimethoprim), and B. bifidum VA07-1AN (resistant to streptomycin). Supplementary Table S2 shows the general features of their genomes. Their size was, in all cases, around 2.2 Mbp but the number of contigs obtained after assembly ranged from 17 to 300. Supplementary Table S3 summarizes some of the key genetic features of the genomes of the sequenced strains. Genes coding for penicillin binding proteins (PBP) were found in all the genomes, although with different numbers and types for the distinct species. Mutations in PBPs encoding-genes known to confer AR were not identified. One gene coding for a D-alanine-D-alanine ligase (Ddl) was detected in each of the strains. In several LAB species, the presence of phenylalanine at the enzyme active site in Ddl is correlated with intrinsic resistance to vancomycin [2]. In addition, in each of the strains, 9-32 genes were classified by the RAST server as belonging to the category “Virulence, Disease, and Defence”, subcategory “Resistance to Antibiotic and Toxic Compounds”. The majority of these genes encoded components dedicated to homeostasis or resistance to heavy metals, such as copper, mercury, and the cobalt-zinc-cadmium triad. Genes encoding elongation factors, efflux pumps, DNA gyrases, and topoisomerases were also included by RAST in this subcategory.
By comparing the genome sequences against the databases CARD, ResFinder, and ARG-ANNOT, no genes known to be involved in AR in L. jensenii VA04-2AN (resistant to trimethoprim), L. paracasei VA02-1AN (resistant to chloramphenicol and vancomycin), and L. reuteri VA24-5 (resistant to trimethoprim and vancomycin) were detected. The only positive correlation between phenotype and genotype was the presence of a conserved phenylalanine (F) residue in the active site of the Ddl ligase, corresponding to amino acid 261 of the Leuconostoc mesenteroides enzyme [2], in the deduced sequence of all vancomycin-resistant (Vmr) strains, while the susceptible (Vms) strains were characterized by the presence of a tyrosine (Y) residue at this position (Figure 2).

Figure 2. Alignment of amino acid sequences around the active site of D-Ala-D-Ala ligases of the five Lactobacillus spp. strains sequenced. Strains with phenylalanine (F) at the enzyme active site (green) show a vancomycin-resistant phenotype, while those having a tyrosine (Y) (pale blue) display a vancomycin-susceptible phenotype.
Genome analysis of L. crispatus VA50-4AN, L. salivarius VA40-10, and B. bifidum VA07-1AN identified no genes known to be involved in aminoglycoside resistance. Therefore, mutations in key genes, such as those coding for the ribosomal S12 protein and others acting on the 16S rRNA molecule, were therefore sought by comparing the DNA and deduced protein sequences from our strains with those in databases. No amino acid differences were observed in the sequences of the ribosomal protein S12 for L. crispatus VA50-4AN and L. salivarius VA40-10 from those belonging to susceptible strains of the same species. Further, alignment of the deduced amino acid sequences for the 16S rRNA guanine(527)-N(7)-methyltranferase (RsmG) proteins of the sequenced strains, showed heterogeneity at several positions between themselves and with respect to sequences in databases. In particular, the RsmG sequence of L. crispatus VA50-4AN showed one amino acid change at position 38 (N→H), while that of B. bifidum VA07-1AN showed three amino acid changes at positions 105 (E→A), 150 (G→D), and 206 (R→G), and that of L. salivarius VA40-10 showed six exclusive amino acid changes at positions 12 (G→E), 67 (D→N), 186 (N→D), 199 (I→V), 208 (Q→K), and 209 (V→I). However, by comparing RsmG sequences from resistant and susceptible strains, none of the changes considered could be associated with streptomycin resistance.
As expected, the genome analysis confirmed the presence of tet(W) in B. bifidum VA07-1AN; this gene was also unequivocally identified by searches in the three AR databases used. The tet(W) gene in B. bifidum VA07-1AN was located in a contig of 76,748 bp. Figure 3 shows the genetic organization of the 40-kbp left extreme of the contig that included the tet(W) gene. The tet(W) sequence of VA07-1AN (1922 bp) was almost identical to that described for Bifidobacterium longum LTBL16 (CP034089.1). Similar tet(W) sequences have also been found in the chromosome of strains belonging to other species such as B. bifidum L22 (NG_048301.1), Lachnospiraceae bacterium KGMB03038 (CP041667.1), and Ruminococcus sp. JE7A12 (CP039381.1); and in plasmids, such as pTZC1 from Cutibacterium acnes TP-CU389 (LC473083.1). Compared to tet(W) in B. longum LTBL16, the tet(W) in B. bifidum VA07-1AN contained an insertion of two extra guanine residues (GG) after nucleotide 731 in the ORF resulting in a frameshift, which produced only a short peptide—289 amino acids long compared to 639 residues for the functional Tet(W). This likely explains the susceptibility of VA07-1AN to tetracycline. The tet(W) gene was flanked by ORFs coding for proteins showing the greatest homology to others from B. longum in the upstream region, and proteins typical of B. bifidum in the downstream region (Supplementary Table S4).
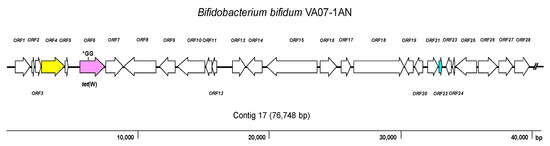
Figure 3. Diagram showing the genetic organization of ORFs in the contig harboring the tet(W) gene of Bifidobacterium bifidum VA07-1AN. Color key: purple, tet(W) gene (the position of the GG insertion disrupting the ORF is indicated); yellow, conjugation-associated gene; pale blue, gene encoding a transcription regulator; white, genes involved in other processes. The broken line symbol indicates the contig extends beyond this point.
The CARD database further identified in the genome of B. bifidum VA07-1AN a single nucleotide polymorphism (SNP) point mutation in the rpsL gene (encoding the ribosomal S12 protein), a variation causing an amino acid substitution (K→R) at position 43 of the protein (Figure 4). This amino acid change has been associated with streptomycin resistance in many species [3].

Figure 4. Alignment of the deduced amino acid sequence of S12 ribosomal proteins encoded by the rpsL gene from streptomycin-resistant (Smr) and -susceptible (Sms) Bifidobacterium bifidum strains. The amino acid replacement K→R at position 43 in the resistant strains is highlighted in pale blue. In bold, the strain of this study (VA07-1AN).
5. Restoration of the Tetracycline Resistance Phenotype in B. Bifidum VA07-1AN
When the susceptibility of B. bifidum VA07-1AN to tetracycline was assayed using the MICE test, colonies growing within the inhibition halo were noted. Identification and typing showed them to be tetracycline-resistant variants of VA07-1AN. After plating on antibiotic-containing and antibiotic-free plates, about 0.6% of the colonies from an overnight culture were found to be tetracycline-resistant. Amplification and sequencing of tet(W) genes from 13 tetracycline-resistant variants showed the addition of one guanine nucleotide in most revertants to the guanine stretch where the two Gs disrupting the ORF had been inserted (as in R-1; Figure 5). Other mutations consisting of both nucleotide insertions and deletions in the vicinity of the stretch of Gs were occasionally seen (as in R-11; Figure 5). In either case, there was a net gain of one nucleotide, which, together with the two Gs that disrupted the tet(W) ORF, created a new codon that opened the reading frame of Tet(W) producing a functional protein that provided tetracycline resistance. The MIC of tetracycline in the tetracycline-resistant variants ranged from 48 to 96 μg mL−1. In contrast, growing the antibiotic-resistant variants in the absence of tetracycline for about 80-100 generations showed no tetracycline-susceptible revertants, demonstrating high stability of the mutations that restored the resistant phenotype.
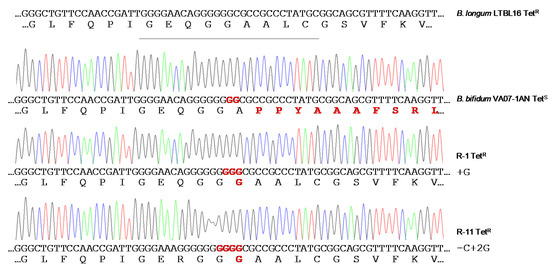
Figure 5. Chromatograms of amplicons of the tet(W) gene from the original tetracycline-susceptible strain B. bifidum VA07-1AN and two representative tetracycline-resistant revertants (R-1 and R-11). Nucleotide sequences and the corresponding deduced amino acid sequences are displayed below each of the chromatograms. DNA and protein differences with canonical sequences of the tet(W) gene from the tetracycline-resistant Bifidobacterium longum LTBL16 strain (on top of the figure) are highlighted in red.
6. Discussion
LAB contribute to the maintenance of vaginal health via the production of substances (mainly organic acids) that acidify the environment and inhibit the development of pathogens [4]. However, there is an increasing concern that LAB may act as reservoirs of AR determinants, from which they could ultimately be transferred to pathogens [5][6]. Indeed, the existence of lactobacilli and bifidobacteria strains resistant to several antibiotics, by either acquiring mutations or exogenous genes, has been repeatedly reported [7][8][9][10][11]. Therefore, during the selection of probiotics, the susceptibility of lactobacilli and bifidobacteria to antibiotics has to be assessed and the absence in the selected strains of transferable AR genes should be assured [12].
Studies reporting lactobacilli and bifidobacteria to be generally susceptible to tetracycline, erythromycin, chloramphenicol, penicillin, ampicillin, clindamycin, quinupristin-dalfopristin, linezolid, and rifampicin have been published over the last 15 years [10][13][14]. In agreement, the phenotypic analysis of the present isolates showed them to be susceptible to these antibiotics, with the exception of ampicillin and chloramphenicol—to which one and three isolates, respectively, were associated with MIC values higher than EFSA’s cut-offs [15]. In contrast, nine isolates showed resistance to one or more aminoglycosides (gentamicin, kanamycin, streptomycin, and neomycin). Resistance to aminoglycosides may occur based on several mechanisms, which include (i) enzymatic modification and inactivation of the antibiotics mediated by aminoglycoside acetyltransferases, nucleotidyltransferases, or phosphotransferases, (ii) increased efflux, (iii) decreased permeability, and (iv) modifications of the 30S ribosomal subunit interfering with the binding of this class of antibiotics [16]. However, most of the MIC values recorded in this study were just one dilution higher than the corresponding cut-off. These small MIC differences might be explained by the normal variation associated with the microdilution assay [17]. Accordingly, none of the aminoglycoside resistance genes searched for by PCR, including the widespread aac(6′)-aph(2″) and aad(E) genes [18], were found in any of the isolates. The genome analysis further discarded the presence of acquired resistances in the sequenced strains, comprising genes and well-characterized mutations involved in aminoglycoside resistance. Given the lack of cytochrome-mediated drug transport, aminoglycoside resistance has been claimed as an intrinsic feature of LAB and other anaerobic bacteria [19]. However, large differences in the MIC values for aminoglycosides even in strains from the same species have been reported in the literature [10][13][14]. The cooperation of other non-specific mechanisms, such as increased membrane impermeability, enhanced activity of unspecific efflux pumps and multi-drug transporters, or the presence of defective cell wall autolytic systems, may further account for differences in MICs between different species and strains [20].
Resistance to the aminoglycoside streptomycin has largely been associated with mutations in chromosomal genes, for example, in rpsL that codes for the ribosomal protein S12 [21], or in rsmG that codes for the 16S rRNA guanine(527)-N(7)-methyltranferase (RsmG) [22]. Comparison of the deduced proteins from streptomycin-susceptible and -resistant strains of the different lactobacilli species analyzed revealed random differences between the RsmG sequences. However, none of them could be consistently associated with streptomycin resistance. In contrast, a mutation in rpsL causing an amino acid change at position 43 (K→R) was observed in B. bifidum VA07-1AN. The same amino acid replacement has been reported in other streptomycin-resistant strains of bifidobacteria [23] and many other species [3], suggesting this to be the most likely explanation for the high resistance to streptomycin shown by VA07-1AN.
Strong resistance to vancomycin is an intrinsic feature in certain Lactobacillus phylogroups and other LAB species such as Leuconostoc spp. [24] caused by an amino acid replacement in the active site of the DdlA ligase (F261Y), as it has been experimentally demonstrated for Leuconostoc mesenteroides [2] and L. reuteri [25].
Although cut-offs for trimethoprim and ciprofloxacin in LAB and bifidobacteria have yet to be defined, strains of most of the present species were associated with quite high MICs. In fact, the resistance of most Lactobacillus species to these antibiotics has been repeatedly reported [10][14]. Folate auxotrophy in lactobacilli is generally accepted as the intrinsic cause of resistance to trimethoprim [26]. Similarly, the reduced affinity of DNA gyrase (GyrA) and topoisomerase IV (ParC) variants for ciprofloxacin and other fluoroquinolones saw in some LAB species has been determined responsible for their insensitivity to this class of antibiotics [27]. Further, the presence of active multidrug efflux systems could contribute to an increase in the MIC for ciprofloxacin in some strains [7]. Since no genes coding for β-lactamases has ever been detected in LAB, non-specific mechanisms, as already discussed for the aminoglycosides, might contribute to the increased MIC of ampicillin in L. crispatus VA50-4AN, as has been reported for L. reuteri [28].
The vaginal lactobacilli in the present study were very susceptible to tetracycline—even though many LAB strains are resistant to it [9][18]. Unexpectedly, PCR analysis detected the presence of tet(W) in the two tetracycline-susceptible B. bifidum strains. This gene has been found to be disseminated among gut-dwelling bacteria of different species from humans and animals [29]. The genome analysis of B. bifidum VA07-1AN showed the gene to contain an insertion of two Gs bases at its center, shifting the ORF and rendering a shorter non-functional peptide. The presence of silent tetracycline resistance genes in bifidobacteria has been reported elsewhere [30][31]. The reactivation of a silent tetracycline resistance phenotype has also been reported for Bifidobacterium animalis subsp. lactis Bb12 [31]. Silent AR genes could, therefore, represent a hazard, even more so when they can be easily reactivated and the restored gene remains stable afterward. Therefore, the use of strains harboring such genes in food and feed systems should be avoided.
References
- Ana Belén Flórez; Ilenia Campedelli; Susana Delgado; Angel Alegría; Elisa Salvetti; Giovanna E. Felis; B. Mayo; Sandra Torriani; Antibiotic Susceptibility Profiles of Dairy Leuconostoc, Analysis of the Genetic Basis of Atypical Resistances and Transfer of Genes In Vitro and in a Food Matrix. PLOS ONE 2016, 11, e0145203, 10.1371/journal.pone.0145203.
- Seon Il-Park; Christopher T. Walsh; D-Alanyl-D-Lactate and D-Alanyl-D-Alanine Synthesis by D-Alanyl-D-Alanine Ligase from Vancomycin-resistantLeuconostoc mesenteroidesEFFECTS OF A PHENYLALANINE 261 TO TYROSINE MUTATION. Journal of Biological Chemistry 1997, 272, 9210-9214, 10.1074/jbc.272.14.9210.
- Munita, J.M.; Arias, C.A; Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 1–37.
- Gilda Tachedjian; Muriel Aldunate; Catriona Bradshaw; R. A. Cone; The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Research in Microbiology 2017, 168, 782-792, 10.1016/j.resmic.2017.04.001.
- Christian Von Wintersdorff; John Penders; Julius M. Van Niekerk; Nathan D. Mills; Snehali Majumder; Lieke B. Van Alphen; Paul H. M. Savelkoul; Petra Wolffs; Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Frontiers in Microbiology 2016, 7, 173, 10.3389/fmicb.2016.00173.
- Thomas U. Berendonk; Célia M. Manaia; Christophe Merlin; Despo Fatta‐Kassinos; Eddie Cytryn; Fiona Walsh; Helmut Bürgmann; Henning Sørum; Madelaine Norström; Marie-Noëlle Pons; Norbert Kreuzinger; Pentti Huovinen; Stefania Stefani; Thomas Schwartz; Veljo Kisand; Fernando Baquero; José Luis Martínez; Tackling antibiotic resistance: the environmental framework. Nature Reviews Genetics 2015, 13, 310-317, 10.1038/nrmicro3439.
- Mohammed Salim Ammor; Ana Belén Flórez; Baltasar Mayo; Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiology 2007, 24, 559-570, 10.1016/j.fm.2006.11.001.
- Sabrina Duranti; Gabriele Andrea Lugli; Leonardo Mancabelli; Francesca Turroni; Christian Milani; Marta Mangifesta; Chiara Ferrario; Rosaria Anzalone; Alice Viappiani; Douwe Van Sinderen; et al.Marco Ventura Prevalence of Antibiotic Resistance Genes among Human Gut-Derived Bifidobacteria. Applied and Environmental Microbiology 2017, 83, e02894-16-16, 10.1128/aem.02894-16.
- Jelena Štšepetova; Heleri Taelma; Imbi Smidt; Pirje Hütt; Eleri Lapp; Evi Aotäht; Reet Mändar; Assessment of phenotypic and genotypic antibiotic susceptibility of vaginalLactobacillussp. Journal of Applied Microbiology 2017, 123, 524-534, 10.1111/jam.13497.
- Ilenia Campedelli; Harsh Mathur; Elisa Salvetti; Siobhan Clarke; M.C. Rea; Sandra Torriani; Paul Ross; Colin Hill; Paul W. O’Toole; Genus-Wide Assessment of Antibiotic Resistance inLactobacillusspp. Applied and Environmental Microbiology 2018, 85, e01738-18, 10.1128/aem.01738-18.
- Vita Rozman; Petra Mohar Lorbeg; Tomaž Accetto; Bojana Bogovič Matijašić; Characterization of antimicrobial resistance in lactobacilli and bifidobacteria used as probiotics or starter cultures based on integration of phenotypic and in silico data. International Journal of Food Microbiology 2020, 314, 108388, 10.1016/j.ijfoodmicro.2019.108388.
- Elisa Salvetti; Paul W. O’Toole; When regulation challenges innovation: The case of the genus Lactobacillus. Trends in Food Science & Technology 2017, 66, 187-194, 10.1016/j.tifs.2017.05.009.
- Susana Delgado; Ana Belén Flórez; Baltasar Mayo; Antibiotic Susceptibility of Lactobacillus and Bifidobacterium Species from the Human Gastrointestinal Tract. Current Microbiology 2005, 50, 202-207, 10.1007/s00284-004-4431-3.
- Hikmate Abriouel; María Del Carmen Casado Muñoz; Leyre Lavilla Lerma; Beatriz Pérez Montoro; W. Bockelmann; Rohtraud Pichner; Jan Kabisch; Gyu-Sung Cho; Charles M.A.P. Franz; Antonio Gálvez; Nabil Benomar; New insights in antibiotic resistance of Lactobacillus species from fermented foods. Food Research International 2015, 78, 465-481, 10.1016/j.foodres.2015.09.016.
- EFSA; Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206.
- Doi, Y.; Wachino, J.I.; Arakawa, Y; Aminoglycoside resistance: The emergence of acquired 16S ribosomal RNA methyltransferases. Infect. Dis. Clin. N. Am. 2016, 30, 523–537.
- Geert R.B. Huys; Klaas D'haene; Margo Cnockaert; Lorenzo Tosi; Morten Danielsen; Ana Belén Flórez; Jaana Mättö; Lars Axelsson; Jenni Korhonen; Sigrid Mayrhofer; Maria Egervärn; Mauro Giacomini; Peter Vandamme; Intra- and Interlaboratory Performances of Two Commercial Antimicrobial Susceptibility Testing Methods for Bifidobacteria and Nonenterococcal Lactic Acid Bacteria. Antimicrobial Agents and Chemotherapy 2010, 54, 2567-2574, 10.1128/aac.00407-10.
- Mohammed Salim Ammor; Ana Belén Flórez; Angela H.A.M. Van Hoek; Clara G. De Los Reyes-Gavilán; Henk J.M. Aarts; Abelardo Margolles; B. Mayo; Molecular Characterization of Intrinsic and Acquired Antibiotic Resistance in Lactic Acid Bacteria and Bifidobacteria. Journal of Molecular Microbiology and Biotechnology 2008, 14, 6-15, 10.1159/000106077.
- Condon, S; Aerobic metabolism of lactic acid bacteria. Irish J. Food Sci. Technol. 1983, 7, 15–25.
- Christopher Elkins; Lisa B. Mullis; Bile-Mediated Aminoglycoside Sensitivity in Lactobacillus Species Likely Results from Increased Membrane Permeability Attributable to Cholic Acid. Applied and Environmental Microbiology 2004, 70, 7200-7209, 10.1128/aem.70.12.7200-7209.2004.
- Andrew P Carter; William M. Clemons; Ditlev Brodersen; Robert J. Morgan-Warren; Brian T. Wimberly; V. Ramakrishnan; Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407, 340-348, 10.1038/35030019.
- Hasan Demirci; Frank V. Murphy; Eileen L. Murphy; Jacqueline L. Connetti; Albert E. Dahlberg; Gerwald Jogl; S T Gregory; Structural Analysis of Base Substitutions in Thermus thermophilus 16S rRNA Conferring Streptomycin Resistance. Antimicrobial Agents and Chemotherapy 2014, 58, 4308-4317, 10.1128/AAC.02857-14.
- Mayumi Kiwaki; Takashi Sato; Antimicrobial susceptibility of Bifidobacterium breve strains and genetic analysis of streptomycin resistance of probiotic B. breve strain Yakult. International Journal of Food Microbiology 2009, 134, 211-215, 10.1016/j.ijfoodmicro.2009.06.011.
- Ellie J. C. Goldstein; Kerin L. Tyrrell; Diane M. Citron; Lactobacillus Species: Taxonomic Complexity and Controversial Susceptibilities. Clinical Infectious Diseases 2015, 60, S98–S107, 10.1093/cid/civ072.
- Jan-Peter Van Pijkeren; Robert A. Britton; High efficiency recombineering in lactic acid bacteria. Nucleic Acids Research 2012, 40, e76, 10.1093/nar/gks147.
- Anne-Kirsten Katla; Hilde Kruse; Gro Johnsen; Hallgeir Herikstad; Antimicrobial susceptibility of starter culture bacteria used in Norwegian dairy products. International Journal of Food Microbiology 2001, 67, 147-152, 10.1016/s0168-1605(00)00522-5.
- William Castro; Maribel Navarro; Christophe Biot; Medicinal potential of ciprofloxacin and its derivatives. Future Medicinal Chemistry 2013, 5, 81-96, 10.4155/fmc.12.181.
- Anna Rosander; Eamonn Connolly; Stefan Roos; Removal of Antibiotic Resistance Gene-Carrying Plasmids from Lactobacillus reuteri ATCC 55730 and Characterization of the Resulting Daughter Strain, L. reuteri DSM 17938. Applied and Environmental Microbiology 2008, 74, 6032-6040, 10.1128/aem.00991-08.
- Thaker, M.; Spanogiannopoulos, P.; Wright, G.D; The tetracycline resistome. Cell. Mol. Life Sci. 2010, 67, 419–431.
- Mohammed Salim Ammor; Ana Belén Flórez; Pablo Álvarez-Martín; Abelardo Margolles; B. Mayo; Analysis of tetracycline resistance tet(W) genes and their flanking sequences in intestinal Bifidobacterium species. Journal of Antimicrobial Chemotherapy 2008, 62, 688-693, 10.1093/jac/dkn280.
- Miguel Gueimonde; Ana Belén Flórez; Angela H. A. M. Van Hoek; Birgitte Stuer-Lauridsen; Per Strøman; Clara G. De Los Reyes-Gavilán; Abelardo Margolles; Genetic Basis of Tetracycline Resistance in Bifidobacterium animalis subsp. lactis. Applied and Environmental Microbiology 2010, 76, 3364-3369, 10.1128/aem.03096-09.




