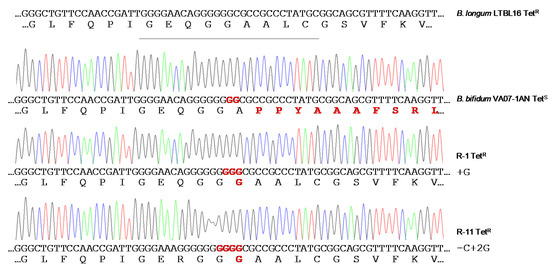
During the selection process of probiotics for vaginal applications, twenty-five lactic acid bacteria (LAB) isolates from human vagina belonging to six different species were tested for antimicrobial resistance by a microdilution method. Gene-specific PCR amplifications proved the strains carry no acquired antibiotic resistance genes, except for a tet(W) gene present in two tetracycline-susceptible Bifidobacterium bifidum strains. Genome analysis of a selected set of strains showed no other acquired resistance determinants. The tet(W) of B. bifidum was inactive by the insertion of two guanine residues in the middle of the gene. Surprisingly, the inactive gene became active and functional very easily, providing resistance to tetracycline and remaining stable afterward. LAB intended to be used in health applications must be free of acquired antimicrobial resistance genes; these could be spread and transferred to human pathogens.
- lactic acid bacteria
- antibiotic resistance
- vaginal microbiota
- genome analysis
- tet(W)
Isolation, Identification and Typing of Vaginal LAB
| Species | Strain | Antibiotic (MIC as µg mL−1) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEN | KAN | STR | NEO | TET | ERY | CLI | CHL | AMP | PEN | VAN | QDA | LIN | TMP | CIP | RIF | ||
| L. crispatus | VA20-32AN | 2 | 16 | 2 | 16 | 2 | 0.06 | 0.25 | 4 | 1 | 0.5 | 0.5 | 1 | 4 | >64 | 16 | 1 |
| VA27-7 | 4 | 32 | 64 | 8 | 1 | 1 | 2 | 8 | 2 | 2 | 1 | 1 | 4 | 32 | 64 | 4 | |
| VA27-9 | 1 | 16 | 2 | 2 | 2 | 0.03 | 0.5 | 4 | 2 | 0.5 | 0.5 | 1 | 4 | 64 | 32 | 2 | |
| VA28-12 | 1 | 16 | 2 | 2 | 2 | 0.06 | 0.5 | 4 | 2 | 0.5 | 0.5 | 2 | 4 | 64 | 32 | 2 | |
| VA32-17 | 2 | 64 | 2 | 8 | 4 | 0.03 | 0.5 | 2 | 1 | 1 | 0.5 | 1 | 2 | >64 | 64 | 8 | |
| VA32-17AN | 4 | 128 | 32 | 4 | 2 | 0.25 | 0.5 | 4 | 1 | 0.5 | 1 | 1 | 2 | 16 | 32 | 4 | |
| VA50-4AN | ≤0.5 | 32 | 1 | 2 | 4 | 0.12 | 0.12 | 4 | 4 | 1 | 0.5 | 1 | 4 | >64 | 32 | 4 | |
| L. jensenii | VA04-1AN | ≤0.5 | 4 | 2 | 1 | 0.25 | ≤0.016 | 0.12 | 4 | 0.25 | 0.12 | 1 | 0.5 | 1 | >64 | 8 | 0.25 |
| VA04-2AN | ≤0.5 | 4 | 4 | 1 | 0.5 | 0.03 | 0.12 | 2 | 0.5 | 1 | 1 | 0.5 | 2 | >64 | 8 | 0.25 | |
| VA15-2AN | ≤0.5 | ≤2 | 1 | ≤0.5 | 1 | ≤0.016 | ≤0.03 | 2 | 0.06 | 0.06 | 0.5 | 0.5 | 0.5 | >64 | 8 | 0.25 | |
| VA16-11 | ≤0.5 | 8 | 1 | 2 | 4 | 0.06 | 0.25 | 4 | 0.06 | ≤0.03 | 2 | 0.5 | 2 | >64 | 16 | 0.5 | |
| Breakpoint (µg mL−1) | 16 | 16 | 16 | - | 4 | 1 | 4 | 4 | 2 | - | 2 | - | - | - | - | - | |
| L. salivarius | VA09-4 | 8 | 64 | 16 | 4 | 2 | 0.25 | 0.25 | 2 | 1 | 0.25 | 128 | 0.25 | 0.5 | ≤0.12 | 1 | 2 |
| VA16-20 | ≤0.5 | 4 | 2 | 0.5 | 1 | 0.06 | 0.06 | 2 | 0.5 | 0.12 | >128 | 0.5 | 0.5 | 0.25 | 0.5 | 0.5 | |
| VA37-13 | ≤0.5 | 4 | ≤0.5 | ≤0.5 | 0.5 | 0.06 | 0.06 | 2 | 0.25 | 0.12 | >128 | 0.5 | 0.5 | 0.25 | ≤0.25 | 0.5 | |
| VA40-10 | 128 | >1024 | >256 | 256 | 2 | 1 | 1 | 4 | 1 | 0.25 | >128 | 1 | 1 | 1 | 4 | 0.5 | |
| VA40-12AN | 4 | 128 | 32 | 4 | 2 | 0.25 | 0.25 | 4 | 0.5 | 0.25 | >128 | 1 | 0.5 | 0.25 | 1 | 1 | |
| VA40-14AN | 4 | 128 | 32 | 4 | 2 | 0.25 | 0.5 | 4 | 0.5 | 0.25 | >128 | 1 | 0.5 | ≤0.12 | 1 | 1 | |
| Breakpoint (µg mL−1) | 16 | 64 | 64 | - | 8 | 1 | 4 | 4 | 4 | - | n.r. | - | - | - | - | - | |
| L. paracasei | VA02-1AN | ≤0.5 | 16 | 8 | 1 | 2 | 0.12 | 0.06 | 8 | 1 | 0.25 | >128 | 1 | 4 | 0.5 | 4 | 0.5 |
| VA24-4 | 1 | 16 | 8 | 4 | 4 | 0.12 | 0.06 | 4 | 0.5 | 0.25 | >128 | 1 | 2 | 0.25 | 4 | 0.5 | |
| VA26-3 | ≤0.5 | 16 | 8 | 2 | 2 | 0.12 | 0.06 | 4 | 1 | 0.25 | >128 | 1 | 2 | 1 | 2 | 0.5 | |
| VA27-8 | 1 | 32 | 16 | 8 | 2 | 0.06 | 0.06 | 8 | 0.5 | 0.25 | >128 | 1 | 4 | 0.25 | 4 | 0.5 | |
| Breakpoint (µg mL−1) | 32 | 64 | 64 | - | 4 | 1 | 4 | 4 | 4 | - | n.r. | - | - | - | - | - | |
| L. reuteri | VA15-3 | ≤0.5 | 4 | 2 | ≤0.5 | 8 | 0.12 | ≤0.03 | 4 | 1 | 2 | >128 | 1 | 2 | >64 | 32 | 0.25 |
| VA24-5 | ≤0.5 | 16 | 4 | ≤0.5 | 16 | 0.06 | ≤0.03 | 4 | 2 | 8 | >128 | 0.5 | 4 | >64 | 32 | 0.25 | |
| Breakpoint (µg mL−1) | 8 | 64 | 64 | - | 32 | 1 | 4 | 4 | 2 | - | n.r. | - | - | - | - | - | |
| B. bifidum | VA07-1AN | 8 | 64 | >256 | 16 | 1 | ≤0.016 | 0.06 | 1 | ≤0.03 | ≤0.03 | 0.5 | 0.5 | 0.5 | 16 | 8 | 2 |
| VA07-2AN | 32 | 64 | >256 | 32 | 1 | ≤0.016 | ≤0.03 | 1 | ≤0.03 | ≤0.03 | 1 | 0.5 | 0.5 | 16 | 8 | 1 | |
| Breakpoint (µg mL−1) | 64 | - | 128 | - | 8 | 1 | 1 | 4 | 2 | - | 2 | - | - | - | - | - | |
Antibiotic Susceptibility
Detection of AR Genes by PCR
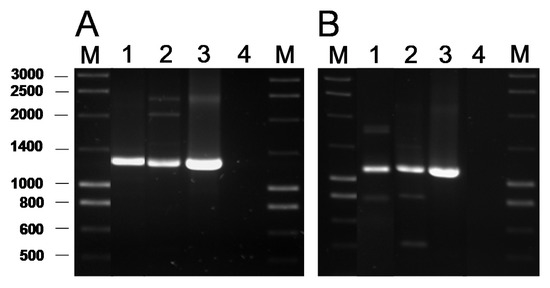
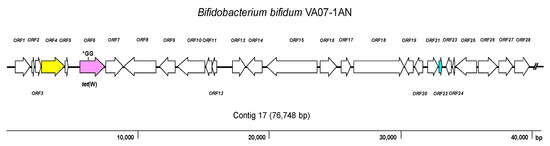
Genome Analysis for AR Genes


Restoration of the Tetracycline Resistance Phenotype in B. Bifidum VA07-1AN
Discussion
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 1–37. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; Mayo, B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef]
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Turroni, F.; Milani, C.; Mangifesta, M.; Ferrario, C.; Anzalone, R.; Viappiani, A.; van Sinderen, D.; et al. Prevalence of antibiotic resistance genes among human gut-derived bifidobacteria. Appl. Environ. Microbiol. 2017, 83, e02894-16. [Google Scholar] [CrossRef]
- Štšepetova, J.; Taelma, H.; Smidt, I.; Hutt, P.; Lapp, E.; Aotäht, E.; Mändar, R. Assessment of phenotypic and genotypic antibiotic susceptibility of vaginal Lactobacillus sp. J. Appl. Microbiol. 2017, 123, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2018, 85, e01738-18. [Google Scholar] [CrossRef] [PubMed]
- Rozman, V.; Lorbeg, P.M.; Accetto, T.; Matijašić, B.B. Characterization of antimicrobial resistance in lactobacilli and bifidobacteria used as probiotics or starter cultures based on integration of phenotypic and in silico data. Int. J. Food. Microbiol. 2020, 314, 108388. [Google Scholar] [CrossRef] [PubMed]
- Salvetti, E.; O’Toole, P.W. When regulation challenges innovation: The case of the genus Lactobacillus. Trends Food Sci. Technol. 2017, 66, 187–194. [Google Scholar] [CrossRef]
- Delgado, S.; Flórez, A.B.; Mayo, B. Antibiotic susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr. Microbiol. 2005, 50, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Abriouel, H.; Casado Munoz, M.D.C.; Lavilla Lerma, L.; Perez Montoro, B.; Bockelmann, W.; Pichner, R.; Kabisch, J.; Cho, G.S.; Franz, C.M.A.P.; Gálvez, A.; et al. New insights in antibiotic resistance of Lactobacillus species from fermented foods. Food Res. Int. 2015, 78, 465–481. [Google Scholar] [CrossRef]
- Doi, Y.; Wachino, J.I.; Arakawa, Y. Aminoglycoside resistance: The emergence of acquired 16S ribosomal RNA methyltransferases. Infect. Dis. Clin. N. Am. 2016, 30, 523–537. [Google Scholar] [CrossRef]
- Huys, G.; D’Haene, K.; Cnockaert, M.; Tosi, L.; Danielsen, M.; Flórez, A.B.; Mättö, J.; Axelsson, L.; Korhonen, J.; Mayrhofer, S.; et al. Intra- and interlaboratory performances of two commercial antimicrobial susceptibility testing methods for bifidobacteria and nonenterococcal lactic acid bacteria. Antimicrob. Agents Chemother. 2010, 54, 2567–2574. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; van Hoek, A.H.; de Los Reyes-Gavilán, C.G.; Aarts, H.J.; Margolles, A.; Mayo, B. Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. J. Mol. Microbiol. Biotechnol. 2008, 14, 6–15. [Google Scholar] [CrossRef]
- Condon, S. Aerobic metabolism of lactic acid bacteria. Irish J. Food Sci. Technol. 1983, 7, 15–25. [Google Scholar]
- Elkins, C.A.; Mullis, L.B. Bile-mediated aminoglycoside sensitivity in Lactobacillus species likely results from increased membrane permeability attributable to cholic acid. Appl. Environ. Microbiol. 2004, 70, 7200–7209. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.P.; Clemons, W.M.; Brodersen, D.E.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Demirci, H.; Murphy, F.V.t.; Murphy, E.L.; Connetti, J.L.; Dahlberg, A.E.; Jogl, G.; Gregory, S.T. Structural analysis of base substitutions in Thermus thermophilus 16S rRNA conferring streptomycin resistance. Antimicrob. Agents Chemother. 2014, 58, 4308–4317. [Google Scholar] [CrossRef] [PubMed]
- Kiwaki, M.; Sato, T. Antimicrobial susceptibility of Bifidobacterium breve strains and genetic analysis of streptomycin resistance of probiotic B. breve strain Yakult. Int. J. Food Microbiol. 2009, 134, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, E.J.; Tyrrell, K.L.; Citron, D.M. Lactobacillus species: Taxonomic complexity and controversial susceptibilities. Clin. Infect. Dis. 2015, 60, S98–S107. [Google Scholar] [CrossRef] [PubMed]
- van Pijkeren, J.P.; Britton, R.A. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 2012, 40, e76. [Google Scholar] [CrossRef]
- Katla, A.K.; Kruse, H.; Johnsen, G.; Herikstad, H. Antimicrobial susceptibility of starter culture bacteria used in Norwegian dairy products. Int. J. Food Microbiol. 2001, 67, 147–152. [Google Scholar] [CrossRef]
- Castro, W.; Navarro, M.; Biot, C. Medicinal potential of ciprofloxacin and its derivatives. Future Med. Chem. 2013, 5, 81–96. [Google Scholar] [CrossRef]
- Rosander, A.; Connolly, E.; Roos, S. Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Appl. Environ. Microbiol. 2008, 74, 6032–6040. [Google Scholar] [CrossRef]
- Thaker, M.; Spanogiannopoulos, P.; Wright, G.D. The tetracycline resistome. Cell. Mol. Life Sci. 2010, 67, 419–431. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; Alvarez-Martín, P.; Margolles, A.; Mayo, B. Analysis of tetracycline resistance tet(W) genes and their flanking sequences in intestinal Bifidobacterium species. J. Antimicrob. Chemother. 2008, 62, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Gueimonde, M.; Flórez, A.B.; van Hoek, A.H.; Stuer-Lauridsen, B.; Strøman, P.; de los Reyes-Gavilán, C.G.; Margolles, A. Genetic basis of tetracycline resistance in Bifidobacterium animalis subsp. lactis. Appl. Environ. Microbiol. 2010, 76, 3364–3369. [Google Scholar] [CrossRef] [PubMed]
This entry is adapted from the peer-reviewed paper 10.3390/ijms21072594
