
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Lidia Ciccone | + 2920 word(s) | 2920 | 2021-04-20 05:35:34 | | | |
| 2 | Conner Chen | Meta information modification | 2920 | 2021-04-25 04:31:42 | | | | |
| 3 | Conner Chen | Meta information modification | 2920 | 2021-04-25 04:32:29 | | |
Video Upload Options
Transthyretin (TTR) is a plasma homotetrameric protein that transports thyroxine and retinol. TTR itself, under pathological conditions, dissociates into partially unfolded monomers that aggregate and form fibrils. Metal ions such as Zn2+, Cu2+, Fe2+, Mn2+ and Ca2+ play a controversial role in the TTR amyloidogenic pathway. TTR is also present in cerebrospinal fluid (CSF), where it behaves as one of the major Aβ-binding-proteins. The interaction between TTR and Aβ is stronger in the presence of high concentrations of Cu2+. Crystals of TTR, soaked in solutions of physiological metals such as Cu2+ and Fe2+, but not Mn2+, Zn2+, Fe3+, Al3+, Ni2+, revealed an unusual conformational change.
1. The TTR structure
Human transthyretin (TTR), or prealbumin, is a homotetrameric protein mainly synthesized in the liver and secreted into the serum. However, a small amount of TTR is also produced in the retina and choroid plexuses of the brain [1]. The acronym TTR encloses the principal protein’s physiological functions: transporter, thyroxine (T4) and retinol in plasma and in cerebrospinal fluid (CSF). While T4 molecules are directly bound to the TTR binding sites, retinol is transported by TTR through its interaction with retinol-binding protein (RBP), which binds orthogonally to T4 [2].
The TTR structure is characterized by four identical 127 amino acid β-sheet sandwich subunits (A, B, A’ and B’) assembled together in a molecular 222 symmetry, Figure 1a. The two dimers (A-B and A’-B’) are oriented to form a central channel that crosses the entire tetramer where T4 binds, Figure 1b [3]. Each monomer folds into a β-sandwich characterized by two β-sheets that consist of four anti-parallel β-strands, DAGH, and CBEF. One short segment of α-helix and a flexible loop connect the β-strands E and F, Figure 1c. The four β-strands DAGH of the four monomers define the channel’s surface of the tetramer, while β-strands CBEF design the external surface.
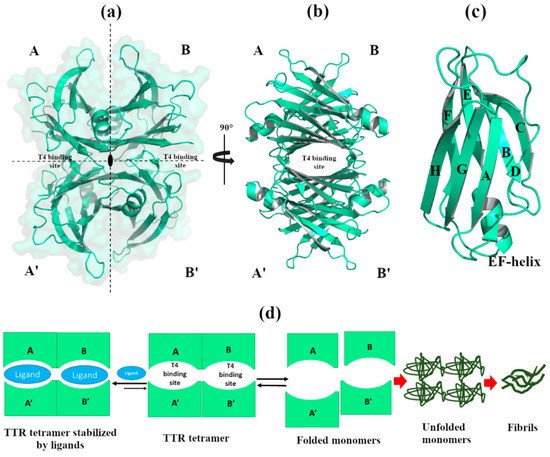
Figure 1. Graphical representation of the human transthyretin (TTR) crystal structure. The figure was created by author using the Protein Data Bank (PDB) id 4TQI [4]). (a) The TTR tetramer is composed by monomers A, A’, B, B’ assembled together around the 222-fold axis. The molecular surface is colored in pale green. (b) TTR tetramer, rotated of 90°, displays the T4 binding sites that cross the entire tetramer. (c) Structural details of monomer A. (d) Schematic representation of amyloidogenic pathway.
2. Effect of Metals in TTR Structure and Function
2.1. Non-Physiological Metals: Cr3+ and Re2+
Starting from a previous study where two halides, iodine and chloride, were investigated for their ability to increase the stability of the TTR tetramer [5], T. Sato et al. screened several metal ions (Cu2+, Zn2+, Ca2+, Co2+, Cd2+, Mn2+, Fe3+ and Cr3+) in order to evaluate their effect on TTR. Among these, only Cr3+ showed a significant reduction in amyloid fibril formation, favoring T4 binding with the tetramer, and stabilizing both wt-TTR and the V30M-TTR variant [6]. The stability of the tetramer was confirmed by calorimetric analyses performed at physiological and acid pH (25 µM TTR samples against 500 µM Cr3+). The X-ray structure of wt-TTR in complex with Cr3+ was solved at 1.8 Å, and the anomalous difference Fourier maps displayed major peaks close to Glu54 (data not deposited) [6]. The authors suggested that Cr3+ can electrostatically neutralize this zone, pushing the T4 to bind TTR [6].
Crystal structures of wt-TTR, in complex with rhenium, were obtained by soaking TTR crystals in cryoprotectant solutions rich in tris-carbonyl derivatives, following a strategy already reported in the literature [7][8]. The initial purpose of this investigation was to study and try to solve the phase problem during the single-wavelength anomalous diffraction (SAD) experiments. Briefly, crystals were obtained by sitting-drop vapor-diffusion method from a reservoir solution of 21% polyethylene glycol 4000 (PEG4K), 0.14 M imidazole malate, pH 6.0 (PDB id:5K1J) as well as from 21% PEG4K, 0.14 M imidazole malate, pH 6.0, 3.6% polyethylene glycol monomethylether (MPEG5K), and 30 mM sodium acetate, pH 5.5 (PDB id 5K1N). For data collection, the first crystal was cryoprotected by soaking for 10 min into cryoprotectant solution composed of 40% of SM2 (12.5% ethylene glycol, 12.5% glycerol, 12.5% 1,2-propanediol, 25% DMSO and 37.5% 1,4-dioxane), 25% PEG 8K and 0.2 mM of rhenium derivative (PDB id:5K1J). The second one was cryoprotected with a solution of 40% SM3 (25% diethylene glycol, 25% ethylene glycol, 25% glycerol, 25% 1,4-dioxane), 25% PEG 8K and 0.5 mM of rhenium compound [7] (PDB id 5K1N). Interestingly, crystals soaked with a low concentration of rhenium derivatives gave structures that diffracted to 1.7 Å resolution, showing a new wt-TTR conformation. Both structures show a Re atom that coordinates with His88 in monomer B and B’, Figure 2a.
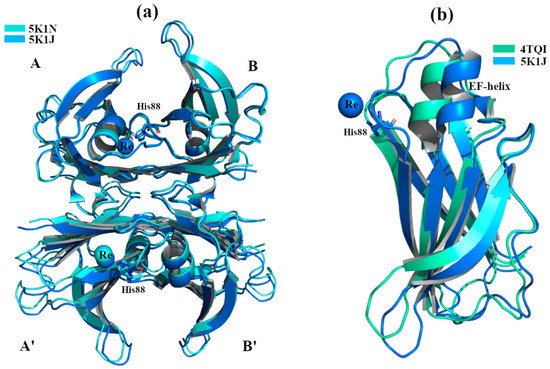
Figure 2. TTR crystal structures in complex with Re2+. The figure was created by author downloading the appropriate PDB codes. (a) Superposition between the two crystals structures soaked with Re2+ under acidic conditions. The common Re2+ binding site is shown. (b) Superposition of monomers B between the standard conformation (PDB 4TQI) versus the new one (5K1J). The major difference is visible around the EF-helix of PDB structure 5K1J that is shifted.
The structural analysis highlighted that the tetramer is well conserved even if the monomers B and B’ in the segment comprised of residues 72-94 (residues 72-90 correspond to EF-helix) are shifted, opening the thyroxin binding site B-B’ while shrinking A-A’ site, Figure 2a,b. This TTR conformation has never been observed before, even if its effect in the central channel is attributable to the well-known negative cooperativity of TTR [9].
Another region of TTR, usually characterized by considerable structural variations, is located between residues 94 to 104 (FG loop). The highest root mean square deviation value calculated on C-α (r.m.s.d.) is that calculated between the new structures and TTR the P31 polymorph (r.m.s.d. 2.74 Å) [7][10]. For more detailed information regarding the structural differences between the TTR-Re crystal complexes and other structures, we refer the reader to the original manuscript [7]. It has been hypothesized that this conformation obtained in the presence of Re, where the EF-helix swings away from the T4 binding site opening the dimer B-B’, may represent the tetramer conformation that is able to interact with the Aβ peptide. This consideration is strengthened by previous studies, where it has been observed that Leu82 (EF-helix) and Leu110 (strand G) are key residues for the interaction between TTR and Aβ [11][12].
2.2. Physiological Metals
2.2.1. Zn2+
Several studies report that Zn2+ binds to TTR, both in vitro and in vivo [13][14][15]. High levels of Zn2+ and Cu2+ ions induce the formation of TTR amyloid deposits in vitro, while chelating agents favor their disruption. In ex-vivo ocular amyloid deposits, from FAP patients with the V30M mutation, elevated concentrations of Zn2+ was found [14]. Starting from this evidence, it was hypothesized that Zn2+, binding with TTR, might induce structural changes triggering its amyloidogenesis process.
X-ray crystal structure analyses of four engineered monomer TTRs (M-TTR, F87M/L110M) [16], in complex with Zn2+ at several concentrations, and at different pHs (pH 7.5, 6.5, 5.5, 4.6), revealed three possible Zn2+ binding sites (ZBS) [17]. Crystals were grown by hanging-drop vapour-diffusion method in a solution composed of 100 mM sodium citrate, 2.0 ammonium sulphate and 200 mM of zinc acetate. The C-α r.m.s.d. among the structures (PDB id: 3DGD, 3GPS, 3GRB, 3GRG) is less than 0.4 Å, indicating that they are very similar to each other, Figure 3a. The binding of Zn2+ with ZBS1 involves the two amino acids Cys10 and His56 and it does not lead to any significant conformational change. This suggests that the allocation of Zn2+ in ZBS1 is physiological and may prevent amyloidogenic character, Figure 3b [17].
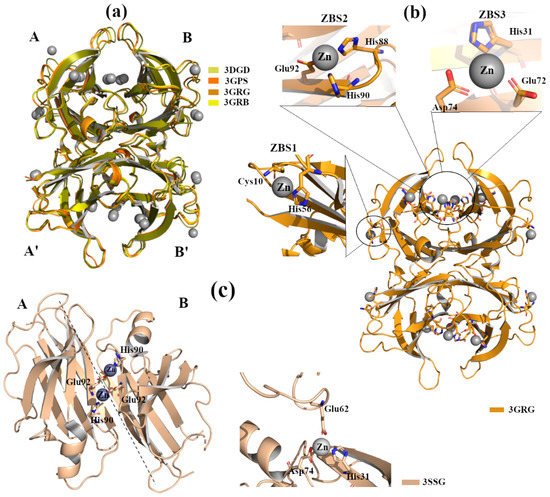
Figure 3. TTR crystal structures in complex with Zn2+. The figure was created by author downloading the appropriate PDB codes. (a) Superposition of the four Zn2+-M-TTR obtained at different pHs (PDB id: 3GRG pH 7.5, 3GRB pH 6.5, 3GPS pH 5.5, 3DGD pH 4.6). Zinc ions are colored in grey. (b) Graphical representation of ZBS1-3 in the M-TTR crystal structure (PDB id: 3GRG). (c) Graphical representation of ZBS1-2 in the L55P-TTR crystal structure (PDB id: 3SSG).
Moreover, the occupation of ZBS1 does not produce any effect on TTR-RBP interaction (the residues of TTR involved in the binding with RBP are Arg21, Val20, Leu82 and Ile-84) [18]. In contrast, in the presence of a higher Zn2+ concentration, the binding of Zn2+ to ZBS2 (Hys88, Hys90 and Glu92) and ZBS3 (Glu72, Asp74 and Hys31) Figure 3b induces slight structural rearrangements around the α-helix at all tested pH. These modifications are comparable with those detected in other TTR structures crystallized without metals but at acidic pH [19]. Contrary to the effect observed for ZBS1, the involvement of ZBS2 and ZBS3 and their corresponding conformational changes affect the interaction of TTR with RBP [17].
TTR L55P is considered as one of the most aggressive amyloidogenic variants that accelerate pathology onset. Structural studies report that apo-TTR L55P, as well as TTR L55P in complex with 2,4-dinitrolphenol (DNP), possess the typical tetrameric structure. The only detected change is local in the monomers, where, due to a disorder of the short edge strand D, an extended loop between strands C and E appears [20][21].
The crystal structure of TTR L55P (PDB id: 3SSG), in complex with Zn2+, does not show significant differences compared to apo-TTR, TTR L55P-DNP and the M-TTR-Zn2+ [22]. TTR L55P, in complex with Zn2+, was grown by hanging-drop vapour-diffusion in presence of 3% w/v PEG 8000, 0.1M cacodylate pH 6.5, 5 mM Zn acetate. This X-ray structure shows two ZBS: ZBS1 is at an intra-dimer site that involves His90 from one monomer and Glu92 from the vicinal monomer, whilst ZBS2 is intra-tetramer, located between His31 and Asp74 from one monomer and Glu62 from symmetric monomer, Figure 3c. No Zn2+ ions are detected around Cys10 and/or His56; this might be related to the proximity of the point mutation L55P. Even if TTR L55P-Zn2+ does not show relevant conformational changes, it is interesting to highlight that Zn2+ binding induces a different tetragonal packing (space group P42212) in the quaternary structure that could represent an ordered intermediate before evolving into an amyloidogenic conformation.
Recently, studies report that TTR can also be considered as an inducible metallopeptidase [15][23]. The TTR catalytic triad is composed of residues His88, His90 and Glu92, and its activation is modulated by bivalent metal ions. It has been demonstrated that, when TTR proteolytic activity is inhibited in vitro by metal chelators, Zn2+ and Mn2+ reestablished their full proteolytic activity, whereas other metals such as Fe2+ and Co2+ only partially reactivated the enzyme. This TTR proteolytic activity agrees with the hypotheses in which TTR behaves as a protease in neurodegenerative diseases such as AD and atherosclerosis. In fact, apoA-I and Aß can be cleaved by TTR, which might affect the onset of atherosclerosis and AD, respectively [24][25].
Despite several studies focusing on understanding whether the interaction between TTR and Zn2+ has a physiological or pathological role, the hypothesis is still debated.
2.2.2. Cu2+, Fe2+ and Mn2+
Biophysical studies employing light scattering and fluorescence spectroscopy demonstrate that Cu2+ binds to TTR; the presence of the metal induces some structural and functional effects on TTR [13]. Binding of Cu2+ provokes a dose-dependent decrease in Trp41 fluorescence intensity, suggesting a local perturbation around this zone. The same tendency was observed when Cu2+ binds TTR in the presence of 1-anilino-8-naphthalene sulfonate (ANS). ANS is a small fluorescent ligand able to bind both T4 binding sites and stabilize the TTR tetrameric structure [26][27]. The observation of a fluorescence perturbation upon Cu2+ binding suggests a local structural variation across the central channel [13]. Interestingly, the same study demonstrated that Cu2+ did not influence the rate of wt-TTR amyloid formation at pH 6.5 or 7.4. This trend was successively confirmed in a urea-induced dissociation experiment. Depending on the tested concentrations, Cu2+ did not show any effect on the tetramer dissociation, but, on the contrary, seemed to stabilize it. In contrast, in the L55P TTR mutant, Cu2+ favors tetramer dissociation and accelerates the process of amyloid formation [13].
As mentioned in the introduction, in contrast with its intrinsic amyloidogenic potential, TTR has a neuroprotective role in AD. TTR binds Aβ, participating in its clearance from the brain [28][29]. It has been hypothesized that metals might play a role in triggering this additional function of TTR.
Structural studies of TTR in complex with different metals confirm that Fe2+, Cu2+ and Mn2+ bind to the protein, Figure 4a. TTR crystals were grown by sitting-drop vapor-diffusion, and the reservoir solution was filled with 21% of PEG4K, 0.14M imidazole malate, pH 6.0 or 21% of PEG4K, 0.14M imidazole malate, pH 6.0, 3.6% MPEG5K, and 30mM sodium acetate, pH 5.5. The cryoprotectant solution was composed of 40% of SM2 (12.5% ethylene glycol, 12.5% glycerol, 12.5% 1,2-propanediol, 25% DMSO and 37.5% 1,4-dioxane) 25% PEG 8K and 30mM of CuCl2, MnCl2 or FeCl2. The X-ray crystal structure analysis of TTR in complex with Mn2+ did not show any significant structural differences. Two possible Mn2+ binding sites were detected around Asp99 and Glu66 in monomer A, and two, Glu66 and Asn98, in monomer B, Figure 4b [30]. In contrast, when wt-TTR crystals were treated with Cu2+, these displayed a similar behavior of those soaked with Fe2+ (Figure 4a) and Re2+ [7][30]. The electron density map suggests that Cu2+ ions are located around His88, His90 and Asp74 for monomer B, and between His90 and Asp74 for monomer A, Figure 4c. A minor pick is detected close to Asp38, Figure 4c.
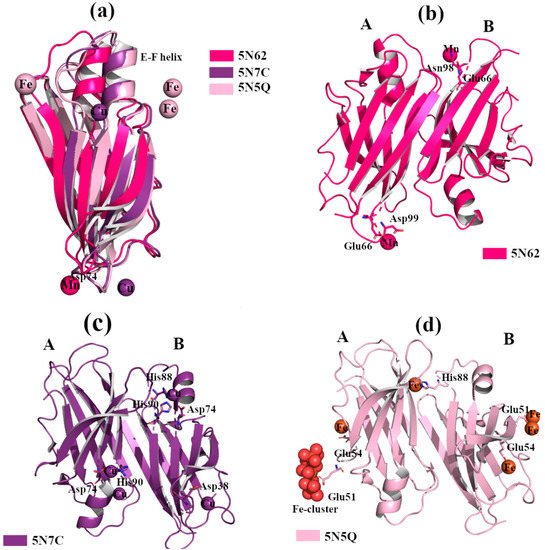
Figure 4. TTR crystal structures in complex with Mn2+, Cu2+ and Fe2+. The figure was created by author downloading the appropriate PDB codes. (a) Superposition of the B monomers of the three TTR crystal structures obtained in presence of Mn2+, Cu2+ and Fe2+, respectively (PDB id 5N62, 5N7C and 5N5Q). The EF-helix of TTR-Mn2+ crystal complex shows the classical conformation, while it is shifted for the other two metals complexes. (b) Graphical representation of the dimer A-B of TTR in complex with Mn2+. (c) Graphical representation of the dimer A-B of TTR in complex with Cu2+. (d) Graphical representation of dimer A-B of TTR in complex with Fe2+.
Thus, Fe2+ and Cu2+, but not Mn2+, induce a conformational change in the wt-TTR tetramer, comparable to that observed in TTR-Re2+ crystal complex [30]. In order to verify if trivalent metals ions also induce conformational changes, several wt-TTR crystals were soaked with Al3+, Gd3+ and Fe3+ following the same protocol. No structural modifications were found.
The binding of Fe2+ with TTR protein was confirmed via the strong anomalous signal registered in the phased anomalous difference Fourier map [31][32]. The highest peak is registered close to Glu51, a second site is located near Glu54 at the entrance of the T4 binding site, and another peak is detected around His88, Figure 4d. As previously seen in the rhenium-TTR structure, the conformational change affects only the β-strands E and F, and the short α-helix connecting them located in monomers B and B’ [30].
Interestingly, the conformational change, induced by Cu2+ and Fe2+, modifies the dimer-dimer interface involving the TTR amino acid sequence, which is implicated in the interaction with Aβ peptides [11][33]. It is known that in the brains of AD patients, the concentration of metals (in particular Cu, Fe and Zn) is altered, and the amount of Cu2+ in Aβ plaque can reach 400 µM [34]. A bio-layer interferometry (BLI) study in solution revealed that a binding affinity between TTR and Aβ1-28 peptide is in nanomolar range when in the presence of Cu2+, thus hinting that the TTR conformational change induced by Cu2+ and Fe2+ might be associated with TTR’s ability to neutralize Aβ [30]. This experimental evidence suggests that the conformational change induced by Cu2+ and Fe2+ is not a structural artefact due to the soaking technique, but is probably related to the neuroprotective role that TTR possesses in the brain. This new conformation of TTR has inspired the design of PROteolysis-Targeting Chimeras (PROTAC) compounds that can induce the “active TTR conformation” favoring both the stabilization of TTR tetramer and the Aβ scavenger [35].
2.2.3. Ca2+
The calcium ion, Ca2+, is one of the most important metals involved in the regulation of cellular signalling pathways and tissue homeostasis. Several studies report that the dysregulation of Ca2+ is a key factor in the triggering of neurodegenerative processes [36]. TTR binds Ca2+ [37], and X-ray studies do not show any relevant structural changes in the wt-TTR tetrameric structure. TTR-Ca2+ crystal complexes were obtained by sitting drop vapour-diffusion from a solution containing 100mM HEPES, 200 mM CaCl2, 28% PEG400, pH 7.5 (PDB id: 4MRB, 200 mM Ca2+ [38]) and 34–40% PEG 400, 400 mM CaCl2, 0.1 M HEPES, pH 7.5 (4N85, 400 mM Ca2+) [39]. The superposition of the two structures reveals that there is a common Ca2+ site between Glu66 and Asp99 of monomer A, while a second site is detected around N-terminus portions in the crystal at higher Ca2+ concentrations, Figure 5a,b. Different variants of TTR have been solved alone or in complex with ligands, in the presence of Ca2+, and in these cases no relevant structural differences were detected [38][40][41].
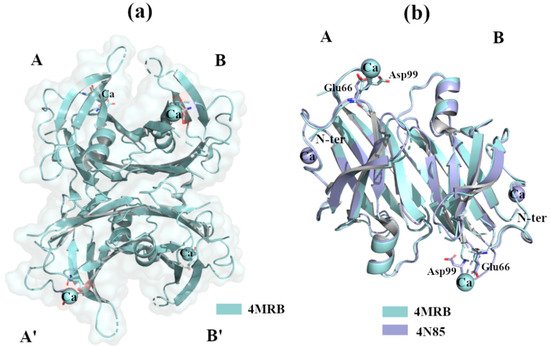
Figure 5. TTR crystal structures in complex with Ca2+. The figure was created by author downloading the appropriate PDB codes. (a) Tetramer representation of TTR in complex with Ca2+. (b) Superposition of the two dimers A-B of TTR in complex with Ca2+.
Recently, studies in solution show that Ca2+ does not modify the environment around tryptophan residues, confirming that it does not induce global structural changes [42]. Interestingly, in the presence of Ca2+, the binding between TTR and ANS decreases, suggesting that the T4 binding sites are less accessible. Deeper analysis confirmed that the fluorescent emission spectra, recorded at 275 nm, did not display any significant structural modifications [42]. However, the same study confirms that Ca2+ increases the rate of fibril formation in the TTR fibril formation assay. This suggests that the dysregulation of Ca2+ ions might have a role in the onset of TTR amyloidosis.
References
- Aldred, A.R.; Brack, C.M.; Schreiber, G. The Cerebral Expression of Plasma Protein Genes in Different Species. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1995, 111, 1–15.
- Hamilton, J.A.; Benson, M.D. Transthyretin: A Review from a Structural Perspective. Cell. Mol. Life Sci. CMLS 2001, 58, 1491–1521.
- Wojtczak, A.; Neumann, P.; Cody, V. Structure of a New Polymorphic Monoclinic Form of Human Transthyretin at 3 Å Resolution Reveals a Mixed Complex between Unliganded and T4-Bound Tetramers of TTR. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001, 57, 957–967.
- Ciccone, L.; Nencetti, S.; Rossello, A.; Tepshi, L.; Stura, E.A.; Orlandini, E. X-ray Crystal Structure and Activity of Fluorenyl-Based Compounds as Transthyretin Fibrillogenesis Inhibitors. J. Enzym. Inhib. Med. Chem. 2016, 31, 824–833.
- Hörnberg, A.; Hultdin, U.W.; Olofsson, A.; Sauer-Eriksson, A.E. The Effect of Iodide and Chloride on Transthyretin Structure and Stability. Biochemistry 2005, 44, 9290–9299.
- Sato, T.; Ando, Y.; Susuki, S.; Mikami, F.; Ikemizu, S.; Nakamura, M.; Suhr, O.; Anraku, M.; Kai, T.; Suico, M.A.; et al. Chromium(III) Ion and Thyroxine Cooperate to Stabilize the Transthyretin Tetramer and Suppress in Vitro Amyloid Fibril Formation. FEBS Lett. 2006, 580, 491–496.
- Ciccone, L.; Policar, C.; Stura, E.A.; Shepard, W. Human TTR Conformation Altered by Rhenium Tris-Carbonyl Derivatives. J. Struct. Biol. 2016, 195, 353–364.
- Ciccone, L.; Vera, L.; Tepshi, L.; Rosalia, L.; Rossello, A.; Stura, E.A. Multicomponent Mixtures for Cryoprotection and Ligand Solubilization. Biotechnol. Rep. 2015, 7, 120–127.
- Neumann, P.; Cody, V.; Wojtczak, A. Structural Basis of Negative Cooperativity in Transthyretin. Acta Biochim. Pol. 2001, 48, 867–875.
- Polsinelli, I.; Nencetti, S.; Shepard, W.; Ciccone, L.; Orlandini, E.; Stura, E.A. A New Crystal Form of Human Transthyretin Obtained with a Curcumin Derived Ligand. J. Struct. Biol. 2016, 194, 8–17.
- Du, J.; Cho, P.Y.; Yang, D.T.; Murphy, R.M. Identification of Beta-Amyloid-Binding Sites on Transthyretin. Protein Eng. Des. Sel. 2012, 25, 337–345.
- Yang, D.T.; Joshi, G.; Cho, P.Y.; Johnson, J.A.; Murphy, R.M. Transthyretin as Both a Sensor and a Scavenger of β-Amyloid Oligomers. Biochemistry 2013, 52, 2849–2861.
- Wilkinson-White, L.E.; Easterbrook-Smith, S.B. Characterization of the Binding of Cu(II) and Zn(II) to Transthyretin: Effects on Amyloid Formation. Biochemistry 2007, 46, 9123–9132.
- Susuki, S.; Ando, Y.; Sato, T.; Nishiyama, M.; Miyata, M.; Suico, M.A.; Shuto, T.; Kai, H. Multi-Elemental Analysis of Serum and Amyloid Fibrils in Familial Amyloid Polyneuropathy Patients. Amyloid 2008, 15, 108–116.
- Liz, M.A.; Leite, S.C.; Juliano, L.; Saraiva, M.J.; Damas, A.M.; Bur, D.; Sousa, M.M. Transthyretin Is a Metallopeptidase with an Inducible Active Site. Biochem. J. 2012, 443, 769–778.
- Jiang, X.; Smith, C.S.; Petrassi, H.M.; Hammarström, P.; White, J.T.; Sacchettini, J.C.; Kelly, J.W. An Engineered Transthyretin Monomer That Is Nonamyloidogenic, Unless It Is Partially Denatured. Biochemistry 2001, 40, 11442–11452.
- Palmieri, L.d.C.; Lima, L.M.T.R.; Freire, J.B.B.; Bleicher, L.; Polikarpov, I.; Almeida, F.C.L.; Foguel, D. Novel Zn2+-Binding Sites in Human Transthyretin: Implications for Amyloidogenesis and Retinol-Binding Protein Recognition. J. Biol. Chem. 2010, 285, 31731–31741.
- Naylor, H.M.; Newcomer, M.E. The Structure of Human Retinol-Binding Protein (RBP) with Its Carrier Protein Transthyretin Reveals an Interaction with the Carboxy Terminus of RBP. Biochemistry 1999, 38, 2647–2653.
- Palaninathan, S.K.; Mohamedmohaideen, N.N.; Snee, W.C.; Kelly, J.W.; Sacchettini, J.C. Structural Insight into PH-Induced Conformational Changes within the Native Human Transthyretin Tetramer. J. Mol. Biol. 2008, 382, 1157–1167.
- Morais-de-Sá, E.; Neto-Silva, R.M.; Pereira, P.J.; Saraiva, M.J.; Damas, A.M. The Binding of 2, 4-Dinitrophenol to Wild-Type and Amyloidogenic Transthyretin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 512–519.
- Cendron, L.; Trovato, A.; Seno, F.; Folli, C.; Alfieri, B.; Zanotti, G.; Berni, R. Amyloidogenic Potential of Transthyretin Variants. J. Biol. Chem. 2009, 284, 25832–25841.
- Castro-Rodrigues, A.F.; Gales, L.; Saraiva, M.J.; Damas, A.M. Structural Insights into a Zinc-Dependent Pathway Leading to Leu55Pro Transthyretin Amyloid Fibrils. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 1035–1044.
- Gouvea, I.E.; Kondo, M.Y.; Assis, D.M.; Alves, F.M.; Liz, M.A.; Juliano, M.A.; Juliano, L. Studies on the Peptidase Activity of Transthyretin (TTR). Biochimie 2013, 95, 215–223.
- Costa, R.; Ferreira-da-Silva, F.; Saraiva, M.J.; Cardoso, I. Transthyretin Protects against A-Beta Peptide Toxicity by Proteolytic Cleavage of the Peptide: A Mechanism Sensitive to the Kunitz Protease Inhibitor. PLoS ONE 2008, 3, e2899.
- Liz, M.A.; Gomes, C.M.; Saraiva, M.J.; Sousa, M.M. ApoA-I Cleaved by Transthyretin Has Reduced Ability to Promote Cholesterol Efflux and Increased Amyloidogenicity. J. Lipid Res. 2007, 48, 2385–2395.
- Cheng, S.-Y.; Pages, R.A.; Saroff, H.A.; Edelhoch, H.; Robbins, J. Analysis of Thyroid Hormone Binding to Human Serum Prealbumin by 8-Anilinonaphthalene-1-Sulfonate Fluorescence. Biochemistry 1977, 16, 3707–3713.
- Lima, L.M.T.R.; Silva, V.d.A.; Palmieri, L.d.C.; Oliveira, M.C.B.R.; Foguel, D.; Polikarpov, I. Identification of a Novel Ligand Binding Motif in the Transthyretin Channel. Bioorg. Med. Chem. 2010, 18, 100–110.
- Alemi, M.; Gaiteiro, C.; Ribeiro, C.A.; Santos, L.M.; Gomes, J.R.; Oliveira, S.M.; Couraud, P.-O.; Weksler, B.; Romero, I.; Saraiva, M.J.; et al. Transthyretin Participates in Beta-Amyloid Transport from the Brain to the Liver- Involvement of the Low-Density Lipoprotein Receptor-Related Protein 1? Sci. Rep. 2016, 6, 1–15.
- Li, X.; Zhang, X.; Ladiwala, A.R.A.; Du, D.; Yadav, J.K.; Tessier, P.M.; Wright, P.E.; Kelly, J.W.; Buxbaum, J.N. Mechanisms of Transthyretin Inhibition of -Amyloid Aggregation In Vitro. J. Neurosci. 2013, 33, 19423–19433.
- Ciccone, L.; Fruchart-Gaillard, C.; Mourier, G.; Savko, M.; Nencetti, S.; Orlandini, E.; Servent, D.; Stura, E.A.; Shepard, W. Copper Mediated Amyloid-β Binding to Transthyretin. Sci. Rep. 2018, 8, 13744.
- Evans, G.; Pettifer, R.F. CHOOCH: A Program for Deriving Anomalous-Scattering Factors from X-Ray Fluorescence Spectra. J. Appl. Cryst. 2001, 34, 82–86.
- Polsinelli, I.; Savko, M.; Rouanet-Mehouas, C.; Ciccone, L.; Nencetti, S.; Orlandini, E.; Stura, E.A.; Shepard, W. Comparison of Helical Scan and Standard Rotation Methods in Single-Crystal X-Ray Data Collection Strategies. J. Synchrotron Radiat. 2017, 24, 42–52.
- Du, J.; Murphy, R.M. Characterization of the Interaction of β-Amyloid with Transthyretin Monomers and Tetramers. Biochemistry 2010, 49, 8276–8289.
- Lovell, M.A.; Robertson, J.D.; Teesdale, W.J.; Campbell, J.L.; Markesbery, W.R. Copper, Iron and Zinc in Alzheimer’s Disease Senile Plaques. J. Neurol. Sci. 1998, 158, 47–52.
- Tonali, N.; Nencetti, S.; Orlandini, E.; Ciccone, L. Application of PROTAC Strategy to TTR-Aβ Protein-Protein Interaction for the Development of Alzheimer’s Disease Drugs. Neural Regen. Res. 2021, 16, 1554.
- Alvarez, J.; Alvarez-Illera, P.; García-Casas, P.; Fonteriz, R.I.; Montero, M. The Role of Ca2+ Signaling in Aging and Neurodegeneration: Insights from Caenorhabditis Elegans Models. Cells 2020, 9, 204.
- Scott, B.J.; Bradwell, A.R. Identification of the Serum Binding Proteins for Iron, Zinc, Cadmium, Nickel, and Calcium. Clin. Chem. 1983, 29, 629–633.
- Mangione, P.P.; Porcari, R.; Gillmore, J.D.; Pucci, P.; Monti, M.; Porcari, M.; Giorgetti, S.; Marchese, L.; Raimondi, S.; Serpell, L.C.; et al. Proteolytic Cleavage of Ser52Pro Variant Transthyretin Triggers Its Amyloid Fibrillogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 1539–1544.
- Yokoyama, T.; Kosaka, Y.; Mizuguchi, M. Crystal Structures of Human Transthyretin Complexed with Glabridin. J. Med. Chem. 2014, 57, 1090–1096.
- Yokoyama, T.; Kosaka, Y.; Mizuguchi, M. Inhibitory Activities of Propolis and Its Promising Component, Caffeic Acid Phenethyl Ester, against Amyloidogenesis of Human Transthyretin. J. Med. Chem. 2014, 57, 8928–8935.
- Murakami, T.; Yokoyama, T.; Mizuguchi, M.; Toné, S.; Takaku, S.; Sango, K.; Nishimura, H.; Watabe, K.; Sunada, Y. A Low Amyloidogenic E61K Transthyretin Mutation May Cause Familial Amyloid Polyneuropathy. J. Neurochem. 2020.
- Wieczorek, E.; Kędracka-Krok, S.; Bystranowska, D.; Ptak, M.; Wiak, K.; Wygralak, Z.; Jankowska, U.; Ożyhar, A. Destabilisation of the Structure of Transthyretin Is Driven by Ca2+. Int. J. Biol. Macromol. 2021, 166, 409–423.




