
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yasuhito Sakuraba | + 5645 word(s) | 5645 | 2021-03-31 04:24:43 | | | |
| 2 | Bruce Ren | -21 word(s) | 5624 | 2021-04-07 04:19:13 | | |
Video Upload Options
Light is the primary regulator of various biological processes during the plant life cycle. Although plants utilize photosynthetically active radiation to generate chemical energy, they possess several photoreceptors that perceive light of specific wavelengths and then induce wavelength-specific responses. Light is also one of the key determinants of the initiation of leaf senescence, the last stage of leaf development. As the leaf photosynthetic activity decreases during the senescence phase, chloroplasts generate a variety of light-mediated retrograde signals to alter the expression of nuclear genes. On the other hand, phytochrome B (phyB)-mediated red-light signaling inhibits the initiation of leaf senescence by repressing the phytochrome interacting factor (PIF)-mediated transcriptional regulatory network involved in leaf senescence.
1. Introduction
Leaf senescence, the final stage of leaf development, is a highly controlled developmental process accompanied by massive transcriptional and metabolic changes that destabilize intracellular organelles and macromolecules and translocate nutrients into developing tissues and storage organs. Numerous studies conducted in the past two decades have greatly expanded our knowledge of the molecular mechanisms underlying the regulation of leaf senescence (reviewed in [1][2][3][4][5]). The initiation of leaf senescence is tightly controlled by endogenous factors, such as the state of phytohormones and other metabolites [5][6] and external stimuli such as drought, high salinity, high temperature, and pathogens [7][8][9][10]. Furthermore, a large number of transcription factors (TFs) involved in the regulation of leaf senescence has been identified in the model plant Arabidopsis (Arabidopsis thaliana) [5][11] and in other plant species [12], uncovering transcriptional regulatory networks that regulate leaf senescence.
Light is the foremost regulator of various biological processes in the plant life cycle. Light characteristics, such as wavelength, fluence rate, and photoperiod, greatly affect plant traits, including growth habit, floral induction, and plant productivity [13]. Plants transform light energy into chemical energy through the process of photosynthesis and also capture light energy using distinct sets of photoreceptors that perceive specific wavelengths of light through the process of light signaling [14]. Light is also a key determinant of the initiation of leaf senescence. In recent years, molecular mechanisms underlying photosynthesis- and light-signaling-mediated regulation of leaf senescence have been uncovered.
2. Regulation of Leaf Senescence by Light via Photosynthesis
Photosynthesis is the process that transforms light energy into chemical energy and thus forms the basis of all life on Earth, ranging from photosynthetic bacteria to higher plants. In plants, photosynthesis takes place in chloroplasts. Since chloroplasts contain approximately 80% of the total leaf nitrogen (N) [15], translocation of N compounds from chloroplasts in older leaves (source organs) to new developing tissues (sink organs) is one of the most crucial events during the process of leaf senescence. In addition, increased expression of senescence-associated genes (SAGs) is promoted by the decline in photosynthetic activity and photosynthesis-related gene expression [16]. Thus, the expression of SAGs during senescence is probably greatly affected by the status of photosynthesis, and the nucleus may obtain the information about the status of photosynthesis through signals produced in chloroplasts. The transport of such retrograde signals from chloroplasts to the nucleus to increase the expression of SAGs is still largely unknown, but several important clues have been found in recent studies.
2.1. Chloroplast-Produced Reactive Oxygen Species (ROS) Affect Leaf Senescence
Reactive oxygen species (ROS) are highly reactive molecules generated by the reduction of ground-state molecular oxygen (3O2) through the acceptance of an electron pair. In plants, the major forms of ROS include superoxide anion (O2−), hydroxyl radical (·OH), hydrogen peroxide (H2O2), and singlet oxygen (1O2). Among these, O2−, ·OH, and H2O2 are produced in most subcellular compartments, while the highly reactive 1O2 is uniquely produced in chloroplasts, which are the primary source of ROS under light conditions [17]. ROS production in chloroplasts is tightly associated with light-dependent photosynthetic reactions. During photosynthesis, 1O2 is produced mainly within photosystem II (PSII) in thylakoid membranes because of the reaction between the triplet state of chlorophyll molecule and 3O2 [17]. On the other hand, the reduction of 3O2 by PSI generates O2−, which is immediately converted into H2O2 on the stromal side of the thylakoid membranes either spontaneously or through the action of superoxide dismutases (SODs) [18]. The production of ROS is drastically increased under unfavorable environmental conditions, such as excess light, drought, and high temperature [19]. In addition, it is well known that the production of ROS, especially H2O2, is significantly increased in leaves during senescence [20], which is accompanied by a significant reduction in the abundance of light harvesting complex II (LHCII) and other photosystem proteins. Consequently, the captured light energy is in excess for the remaining photosynthetic units, leading to the production of a large amount of ROS even under moderate light conditions. In addition, the dismantling of LHCII subunits releases a large number of chlorophyll molecules and their degradation products. It is well known that several chlorophyll biosynthesis intermediates, including protoporphyrin IX and protochlorophyllide, act as photosensitizers that produce high amounts of 1O2 under light conditions [21][22]. Similarly, several chlorophyll degradation products, such as 7-hydroxymethyl chlorophyll a, pheophorbide a, and red chlorophyll catabolite, also act as photosensitizers [23][24][25] (Figure 1).
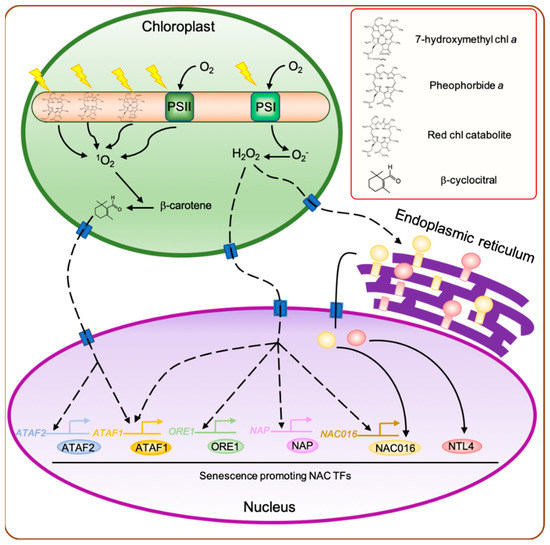
Figure 1. Regulatory network of leaf senescence induced by chloroplast-derived retrograde signaling. During the senescence phase, the abundance of photosystem proteins decreases significantly. Consequently, the captured light energy exceeds the amount needed by the remaining photosynthetic units, leading to the production of a large amount of O2− by photosystem I (PSI) and 1O2 by PSII. In addition, the dismantling of light harvesting complex II (LHCII) proteins releases a large number of chlorophyll molecules and their degradation products, such as 7-hydroxymethyl chlorophyll a, pheophorbide a, and red chlorophyll catabolite, which act as photosensitizers, producing high amounts of 1O2 in the presence of light. The 1O2 generated in chloroplasts promotes the cleavage of β-carotene to produce β-cyclocitral, which then acts as a retrograde signal between the chloroplast and nucleus to activate the expression of 1O2-responsive genes, including senescence-associated Arabidopsis thaliana activating factor (ATAF) subfamily no apical meristem/ATAF1,2/cup-shaped cotyledon (NAC) genes, ATAF1 and ATAF2. O2− generated by PSI is rapidly dismutated into H2O2, which acts as a signaling molecule to alter the expression of H2O2-responsive genes, including ATAF1, ORESARA1 (ORE1), NAC-like, activated by AP3/PI (NAP), and ANAC016. In addition, H2O2 generated in the chloroplasts also triggers the translocation of membrane-bound NAC transcription factors (TFs), such as ANAC016 and NAC with transmebrane motif 1-LIKE4 (NTL4), from the endoplasmic reticulum to the nucleus. Dashed arrowheads indicate the indirect activation of genes. chl, chlorophyll.
For many years, it has been proposed that ROS generated in chloroplasts are involved in the promotion of leaf senescence (reviewed in [26][27]). Among these ROS, H2O2 plays a crucial role in the regulation of leaf senescence. Since H2O2 can easily cross cellular membranes [28], it can be transmitted as a signaling molecule from the chloroplast to the nucleus. Transcriptome analyses of Arabidopsis revealed that several genes upregulated during leaf senescence are also upregulated by H2O2 treatment [29][30]; for example, no apical meristem/ATAF1,2/cup-shaped cotyledon (NAC) TF-encoding genes, such as Arabidopsis thaliana activating factor1 (ATAF1)/ANAC002 [31], ANAC016 [32], NAC-like, activated by AP3/PI (NAP)/ANAC029 [33], JUNGBRUNNEN1 (JUB1)/ANAC042 [34], and ORESARA1 (ORE1)/ANAC092 [35], which have been characterized as key regulators of leaf senescence (Figure 1). Among these, JUB1 acts as a negative regulator of leaf senescence, while the other four NAC genes promote leaf senescence.
H2O2 is also one of the triggers that induce the translocation of membrane-bound NAC TFs from the endoplasmic reticulum (ER) membrane to the nucleus. Several membrane-bound NAC TFs are activated by H2O2 generated in mitochondria under environmental stresses [36][37]. The induction of nuclear gene expression by treatment with methyl viologen, which accepts electrons from PSI via ferredoxin and leads to the production of a large amount of O2− in chloroplasts, is strongly inhibited in the knockout mutant of ANAC017, an Arabidopsis membrane-bound NAC TF gene [38], indicating that H2O2 generated in chloroplasts is also important for the activation of membrane-bound NAC TFs. In Arabidopsis and rice (Oryza sativa L.), several membrane-bound NAC TFs including ANAC016 [32], NAC WITH TRANSMEMBRANE MOTIF 1-LIKE4 (NTL4)/ANAC053 [39], and ONAC054 [40] have been shown to promote the initiation of leaf senescence. Thus, H2O2 generated in chloroplasts during the senescence phase may modulate the gene expression partially via the activation of senescence-associated membrane-bound NAC TFs (Figure 1).
O2− also modulates the expression of SAGs. In Arabidopsis seedlings treated with methyl viologen, an O2−-specific propagator, several genes encoding senescence-associated TFs including ATAF1, WRKY6 [41], and WRKY22 [42] were strongly upregulated [43]. WRKY6 and WRKY22 antagonistically regulate leaf senescence; WRKY6 acts as a negative regulator, while WRKY22 acts as an enhancer of leaf senescence. In contrast with H2O2, O2− is a short-lived ROS and cannot cross the chloroplast membrane [44]. Thus, O2− probably generated in chloroplasts cannot serve as a signaling molecule.
Compared with H2O2 and O2−, 1O2 is a highly reactive molecule and consequently a more potent oxidizing agent than the other ROS [45]. In leaf tissues, in particular, 1O2 is required for most of the lipid peroxidation reactions. Lipid peroxidation promotes the generation of free radicals, which accelerate senescence [46]. 1O2 also gives rise to a signal that affects the expression of nuclear genes, similar to the other two ROS. Op den Camp et al. (2003) investigated the effect of 1O2 on nuclear gene expression using Arabidopsis flu mutant, which shows greater accumulation of protochlorophyllide than the wild type. Since protochlorophyllide acts as a photosensitizer, the flu mutant generates a large amount of 1O2 when transferred from dark to light conditions [22]. In dark-grown flu mutant seedlings, incubation under light for 2 h significantly upregulated the expression of several stress-responsive genes including ABSCISIC ACID (ABA) INSENSITIVE 1 (ABI1), which encodes a protein phosphatase that acts as a negative regulator of ABA signaling [47], and 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE 4 (ACO4), which encodes an ethylene biosynthesis enzyme [48]. In addition to their roles in abiotic stress response, ABA and ethylene are also known to promote leaf senescence [49]. Thus, 1O2 may affect the transcriptional regulatory networks of leaf senescence by modulating the expression of stress response and leaf-senescence-related genes.
2.2. State of Photosystem Proteins Determines the Initiation of Leaf Senescence
Several studies indicate that the state of photosystem proteins affects the initiation of leaf senescence. Chlorophyllide a oxygenase (CAO) is a chlorophyll biosynthesis enzyme that catalyzes two reactions: the conversion of chlorophyll a into 7-hydroxymethyl chlorophyll a and the subsequent transformation of 7-hydroxymethyl chlorophyll a into chlorophyll b [50]. The N-terminal domain of CAO proteins, which is required for their destabilization, is conserved among higher plants [51]. Transgenic Arabidopsis plants overexpressing N-terminal domain-truncated CAO overaccumulated chlorophyll b and stayed green during the senescence phase under both light and dark conditions [52]. Moreover, in these transgenic plants, the abundance of chlorophyll b in the antenna proteins of PSI and PSII increased significantly, and chlorophyll b was also incorporated into CP43, a PSII core protein, which otherwise binds only to chlorophyll a [53]. One of the possible explanations for the stay-green phenotype of these transgenic plants is the stabilization of photosystem proteins due to the increased incorporation of chlorophyll b in photosystem core complexes. Similar to the CAO overexpression lines, Arabidopsis and rice knockout mutants of NON-YELLOW COLORING1 (NYC1), which encodes chlorophyll b reductase, also showed the stay-green phenotype with highly retaining of antenna proteins of photosystems, as well as the structure of grana thylakoid, during senescence [54][55]. Rice delayed yellowing1 (dye1) mutants, in which the pigment-binding function of one of the LHCI subunits (Lhca4) was impaired, exhibited prolonged greenness during the senescence phase [56]. In dye1 mutant plants, although the accumulation of other Lhca proteins (Lhca1, Lhca2, and Lhca3) was similar to that in the wild type, Lhcb1 (a subunit of LHCII) showed high accumulation [56]. Since the chlorophyll b content of the dye1 mutant increased because of the increase in the chlorophyll b-binding capacity of LHCII proteins, this result also supports the hypothesis that the abundance of chlorophyll b is positively correlated with the stability of PSI and PSII during senescence. Additionally, a large number of SAGs were differentially expressed between CAO-overexpressing and wild-type Arabidopsis plants [52]. Thus, the stabilization of photosystem proteins, partially by chlorophyll b, is one of the key factors affecting the abundance of signaling molecules required for the induction of leaf-senescence-related transcriptional-regulatory networks.
2.3. Potential Role of Chlorophyll and Caroteinoid Degradation Products as Retrograde Signaling Molecules in the Regulation of Leaf Senescence
Chlorophyll biosynthesis and degradation intermediates have the potential to produce ROS as photosensitizers, as described above. However, according to several other studies, some chlorophyll intermediates can mediate retrograde signaling from the chloroplasts to the nucleus. It has been shown that Mg-protoporphyrin IX (Mg-proto IX) can act as a negative regulator of nuclear genes associated with photosynthesis. Exogenous application of Mg-proto IX to Arabidopsis protoplasts has been shown to greatly inhibit the expression of Lhcb [57]. In addition, Mg-proto IX can bind to heat-shock protein 90 (HSP90) to decrease its ATPase activity, which subsequently decreases the expression of genes associated with photosynthesis, probably via the regulation of LONG HYPOCOTYL5 (HY5) [58]. Considering the function of Mg-proto IX, it is also probable that chlorophyll degradation intermediates, such as 7-hydroxymethyl chlorophyll a, pheophytin a, and pheophorbide a, act as signaling molecules. Indeed, several genes associated with chlorophyll degradation were upregulated in transgenic Arabidopsis plants overexpressing PHEOPYTINASE (PPH) [59], which encodes a chlorophyll degradation enzyme that catalyzes the conversion of pheophytin a into pheophorbide a [60]. Similarly, STAY-GREEN1 (SGR1), a chlorophyll degradation enzyme that converts chlorophyll a into pheophytin a [61], also affects the expression of genes associated with leaf senescence. For example, when SGR1 was overexpressed via the dexamethasone (DEX)-inducible system, genes associated with chlorophyll degradation, such as PPH, SGR2 [61], NYC1 [55], and PAO [24], and other SAGs, such as ORE1, were significantly upregulated [62]. These results indicate that the accumulation of chlorophyll degradation intermediates or changes in the activity of chlorophyll degradation enzymes may lead to retrograde signaling between the chloroplast and nucleus to activate the transcriptional regulatory networks involved in leaf senescence. The other possibility is that chlorophyll degradation intermediates themselves act as signaling molecules.
Ramel et al. (2012) revealed that in Arabidopsis, high light-induced 1O2 promotes the cleavage of β-carotene, and the resulting breakdown products including β-cyclocitral mediate 1O2-related retrograde signaling to modulate the expression of genes associated with photooxidative stress [63]. Among these genes, four NAC genes, ATAF1, ANAC032, ATAF2/ANAC081, and ANAC102, which encode a senescence-promoting NAC TF [31][64], are strongly upregulated by both β-cyclocitral and high light intensity [65]. Given the elevated level of 1O2 in chloroplasts during senescence phase, β-cyclocitral-mediated retrograde signaling may play a role in the regulation of leaf senescence
3. Regulation of Leaf Senescence by Light Signaling
In recent years, numerous studies have been conducted to understand the relationship between light signaling and leaf senescence, and their results have expanded our knowledge of the role of light signaling in the initiation of leaf senescence. In Section 3.1, the current knowledge of the functions of plant photoreceptors is briefly summarized as a background to the light-signaling-mediated regulation of leaf senescence.
3.1. Role of Photoreceptors in Plants
Plant growth and development are optimized through the perception of light across the spectral range of 300–800 nm [14]. Within this spectrum, light wavelengths ranging from 400 to 700 nm, which together comprise photosynthetically active radiation [66], are used for photosynthesis. On the other hand, plants have distinct sets of photoreceptors that together enable the perception of light across a broad spectrum, ranging from ultraviolet-B (UV-B, 280–315 nm) to far-red (700–800 nm), and the induction of wavelength-specific responses. Cryptochrome is one of the blue-light receptors that perceives blue light (approximately 380–500 nm) and UV-A light (approximately 320–380 nm) and is involved in the regulation of various biological phenomenon including the circadian clock, flowering, and photomorphogenesis [67]. Another type of blue-light photoreceptor, phototropin, regulates chloroplast movement, stomatal opening, and phototropic curvature [67]. Furthermore, three LOV/F-box/Kelch-repeat proteins, namely ZEITLUPE (ZTL), FLAVIN-BINDING KELCH REPEAT, F-BOX1 (FKF1), and LOV KELCH REPEAT PROTEIN2 (LKP2), have also been identified as blue-light receptors and are involved in the regulation of circadian rhythms and photoperiodic flowering [66]. UV RESISTANCE LOCUS 8 (UVR8) perceives UV-B light and regulates UV-B-light-specific responses such as cotyledon opening and flavonoid synthesis [68]. Phytochrome is the sole photoreactor that perceives red light (approximately 600–700 nm) and far-red light and regulates a variety of physiological processes including flowering, seed germination, seedling de-etiolation, and shade avoidance response [69]. The model plant Arabidopsis contains five phytochromes, namely phyA, phyB, phyC, phyD, and phyE [70]. These phytochromes can be divided into two groups, according to their primary photosensory activities and physiological roles. Among these, phyB–phyE are involved in the red/far-red low-fluence response via the reversible transition between the red-light-absorbing (biologically inactive) Pr form and the far-red-light absorbing (biological active) Pfr form; only the Pfr form can move from the cytosol to the nucleus to activate the red-light signaling pathway. Among these four phytochromes, the abundance of phyB is far greater than those of the other three phytochromes. Unlike these four phytochromes, phyA plays an important in the far-red-light response [71]. Thus, these photoreceptors differentially regulate a variety of light-responsive events.
When these photoreceptors are activated by a specific wavelength of light, they move into the nucleus and modulate the activity of TFs specific for each light signaling cascade. For instance, under red-light illumination, the biologically active Pfr form moves into the nucleus, and it interacts with phytochrome interacting factors (PIFs), which are helix–loop–helix (bHLH) TFs and acts as a negative regulator of red light signaling [72]. Similarly, photoactivated cryptochromes move into the nucleus to interact with CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX (CIB) TFs to negatively regulate blue-light signaling [73]. A basic leucine zipper (bZIP)-type transcription factor, HY5, and its functional homolog, HY5 HOMOLOG (HYH), are also key regulators of light signaling. The activity of HY5 and HYH is regulated by the CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1)–SUPPRESSOR OF PHYA1 (SPA1) complex in the nucleus via the E3 ligase activity of COP1 [74]. Importantly, three different light components (red light, far-red light, and blue light) affect COP1–SPA1-mediated degradation of HY5 [74]. Thus, HY5 and HYH act as key regulators of light signaling by integrating the information from multiple light signaling pathways. Recent studies revealed some of the molecular mechanisms underlying red-, far-red-, and blue-light-mediated regulation of leaf senescence.
3.2. Red-Light Signaling-Mediated Regulation of Leaf Senescence in the Model Plant Arabidopsis
Red light has long been considered to delay leaf senescence. In 1970s and 1980s, the effect of red light on the initiation of leaf senescence was extensively investigated and was shown to retard chlorophyll degradation in the leaves of barley (Hordeum vulgare L.) [75][76], tomato (Solanum lycopersicum L.) [77], cucumber (Cucumis sativus L.) [77], and mustard (Sinapis alba L.) [78], indicating that red light delays the initiation of leaf senescence.
In contrast to previous research, molecular mechanisms underlying red-light signaling-mediated promotion of leaf senescence have been revealed in the last decade, especially in Arabidopsis. During dark-induced leaf senescence, yellowing of cotyledons in the A. thaliana ecotype Columbia (Col-0; wild type) was strongly inhibited by intermittent red-light pulse; however, this delayed leaf senescence phenotype was diminished by far-red-light pulse [79]. In addition, phyB mutants exhibited insensitivity to red-light pulse-induced retardation of leaf yellowing, indicating that phyB acts as the main photoreceptor for red-light signaling-mediated promotion of leaf senescence [79]. Among the four PIFs (PIF1, PIF3, PIF4, and PIF5) that are regulated by the active Pfr form of phyB, PIF4 and PIF5 positively regulate both age-dependent and dark-induced leaf senescence [79] (Figure 2).
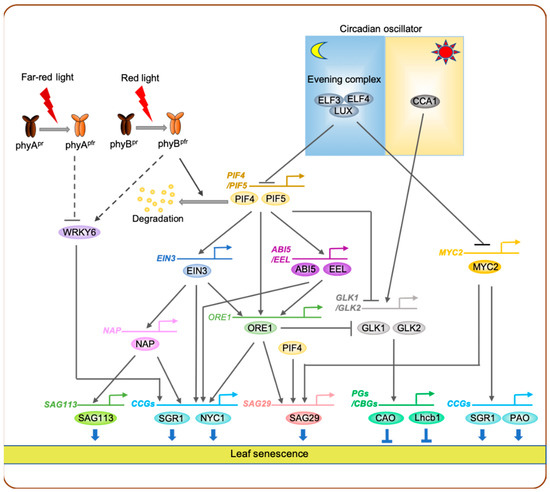
Figure 2. Transcriptional regulatory network of leaf senescence regulated by light signaling. Under red light, PIF4 and PIF5 are degraded through a phyB-mediated proteasomal degradation pathway, leading to the inactivation of PIF-dependent promotion of leaf senescence. In darkness, however, phyB exists in the inactive Pr state, which allows PIF4 and PIF5 to activate the expression of ETHYLENE INSENSITIVE3 (EIN3), ABSCISIC ACID INSENSITIVE 5 (ABI5), and ENHANCED EM LEVEL (EEL). Then PIFs, together with EIN3, ABI5, and EEL, form a feedforward loop to activate the expression of ORE1. Among the proteins that act downstream of PIF4 and PIF5, ORE1 is involved in feedforward loops that activate genes, encoding chlorophyll degradation enzymes, repressing GOLDEN2-LIKE (GLK) genes, which encode transcriptional activators of photosynthesis and chlorophyll biosynthesis-related genes. In the circadian oscillator, the EARLY FLOWERING3 (ELF3)–ELF4–LUX evening complex (EC) represses the expression of PIF4 and PIF5 as well as that of MYC2, which encodes a key TF involved in jasmonate signaling, while CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), a core component of the circadian oscillator, activates the expression of GLK genes and represses the expression of ORE1. WRKY6, which encodes a senescence-associated WRKY TF, is negatively and positively regulated by phyA- and phyB-mediated signaling pathways, respectively. Solid lines indicate direct regulation, while dashed lines indicate indirect regulation. CCGs, chlorophyll catabolic genes; PGs, photosynthesis genes; CBGs, chlorophyll biosynthetic genes.
A number of downstream target genes of PIF4 and PIF5 involved in leaf senescence pathways have been identified to date. During dark-induced leaf senescence, PIF4 and PIF5 directly activate the transcription of ORE1, which encodes a senescence-associated NAC TF and is considered a key promoter of leaf senescence in Arabidopsis [35]. Furthermore, both PIF4 and PIF5 mediate the signaling of two senescence-promoting phytohormones, ethylene and ABA [80][81], by directly activating the expression of ETHYLENE INSENSITIVE 3 (EIN3), which encodes a key ethylene signaling TF [82], and ABI5 and ENHANCED EM LEVEL (EEL), which are two sister genes encoding bZIP-type ABA signaling TFs [83]. Moreover, EIN3, ABI5, and EEL were also shown to directly activate ORE1 expression. Thus, PIF4, PIF5, EIN3, ABI5, and EEL form multiple coherent feedforward loops to activate ORE1 expression [79] (Figure 2). Both PIF4 and PIF5 were also shown to directly activate the transcription of genes encoding chlorophyll catabolic enzymes. PIF4 and PIF5 directly bind to the promoter of SGR1 to activate its expression [84][85]. In addition, PIF5 also activates the transcription of NYC1 by binding to its promoter [85]. Moreover, ABI5, EEL, ORE1, and EIN3 also directly activate SGR1 and NYC1 expression [79][86]. Thus, PIF4, PIF5, and their downstream senescence-associated TFs form coherent feedforward loops to activate genes associated with chlorophyll degradation during leaf senescence (Figure 2). Such coherent feedforward loops regulate many biological processes in plants, such as chlorophyll biosynthesis, floral induction, and leaf senescence [35][87][88], and are thought to increase the robustness of transcriptional regulatory networks [89].
GOLDEN2-LIKE 1 (GLK1) and GLK2, which encode GARP-type MYC TFs that directly activate genes encoding photosystem proteins and chlorophyll biosynthesis enzymes [90], have also been identified as downstream targets of PIF TFs during leaf senescence. In darkness, PIF4 and PIF5 protein levels increase because of the reduction in the activity of phytochromes [79], leading to decreased expression levels of GLKs [84]. Consequently, phenomena such as photosynthesis and chlorophyll biosynthesis are downregulated during dark-induced leaf senescence. PIF TFs have also been shown to directly activate or repress other known SAGs, including JUB1/ANAC042 [91] and SENESCENCE ASSOCIATED GENE 29 (SAG29) [92], although the significance of these transcriptional cascades in leaf senescence has not yet been validated. Taken together, in Arabidopsis, phyB-mediated red-light signaling inhibits the initiation of leaf senescence via multiple regulatory cascades composed of many SAGs (Figure 2).
Recently, in Arabidopsis, Kim et al. (2019) reported that phyB and PIFs also mediate high-temperature-induced leaf senescence. While the light-activated Pfr form of phyB is inactivated in darkness at 20 °C, it is inactivated more rapidly at 28 °C [93]. This inactivation of phyB at high temperature leads to an increase in PIF4 protein levels, which activates ORE1 expression directly or indirectly through ABA and ethylene signaling, thus promoting leaf senescence at high temperature [93]. Thus, the phyB–PIF signaling module also integrates temperature signaling to regulate leaf senescence.
3.3. Red-Light Signaling-Mediated Regulation of Leaf Senescence in Crops
While the mechanisms underlying phyB-mediated red-light signaling in the regulation of leaf senescence have been intensively studied in Arabidopsis, information available in other plant species is limited. Piao et al. (2015) reported that similar to Arabidopsis phyB mutants, rice phyB (OsphyB) T-DNA insertion knockout mutants exhibited accelerated leaf yellowing during dark-induced senescence [94], indicating that Arabidopsis and rice phyB homologs exhibit similar roles in the regulation of leaf senescence. Among the PIF TFs in rice, PIF-LIKE1 (OsPIL1; also known as OsPIL13) shows high sequence similarity with Arabidopsis PIF4 and PIF5 [95]. However, unlike the delayed leaf yellowing phenotype of Arabidopsis pif4 and pif5 mutants, leaves of the ospil1 knockout mutant turned yellow much faster than wild-type leaves [96], suggesting that OsPIL1 acts as a negative regulator of leaf senescence in rice. Interestingly, transgenic Arabidopsis plants overexpressing OsPIL1 exhibited an accelerated leaf yellowing phenotype during dark-induced leaf senescence, similar to Arabidopsis pif4 and pif5 mutants [96], indicating that OsPIL1 and Arabidopsis PIF4 and PIF5 play similar roles in the regulation of leaf senescence, at least in Arabidopsis. Probably, the differences in their downstream cascades between Arabidopsis and rice modified their effects on the induction of leaf senescence. In a similar case, Arabidopsis EARLY FLOWERING3 (ELF3) acts as a negative regulator of leaf senescence [79], while OsELF3 acts as a positive regulator since oself3 knockout mutants exhibited delayed leaf yellowing during both natural and dark-induced leaf senescence [97], in contrast with Arabidopsis elf3 mutant [79]. Since ELF3 downregulates PIF4 and PIF5 expression in Arabidopsis [79], OsPIL1 may be one of the key downstream target genes of OsELF3 in the leaf senescence pathway, and may explain, at least partially, the opposite effects of ELF3 homologs on leaf senescence initiation in Arabidopsis and rice.
In tomato, SlPIF4 was found to exhibit similar functions as Arabidopsis PIF4 and PIF5 and has been shown to affect plant size, flowering, auxin level, and thermomorphogenesis [98]. In addition, silencing of SlPIF4 via RNA interference (RNAi) delayed leaf yellowing during the senescence phase and downregulated several SAGs, such as SlSGR1, SlSAG12, and SlORE1 [98]. These findings suggest that SlPIF4 promotes leaf senescence by modulating downstream transcriptional cascades, similar to Arabidopsis PIF4 and PIF5. Moreover, similar to transgenic Arabidopsis lines overexpressing OsPIL1, those overexpressing maize PIF4 (ZmPIF4) also exhibited accelerated leaf-yellowing phenotype during the senescence phase [99]. Phenotypic characterization of ZmPIF4 overexpression lines and zmpif4 knockout mutants is necessary to further understand the role of ZmPIF4 in the regulation of leaf senescence.
3.4. Red-Light Signaling-Mediated Regulation of Leaf Senescence Induced by Nutrient Deficiency
While phyB-mediated red-light signaling regulates dark-induced leaf senescence, as described above, this mechanism has also been shown to affect leaf senescence under light conditions. Detached leaves of osphyB knockout mutants turned yellow much faster than wild-type leaves when they were incubated in nutrient-free liquid medium under continuous light conditions [94]. In addition, the early-yellowing phenotype of osphyB mutant leaves was recovered by the supplementation of N compounds, such as potassium nitrate (KNO3) and ammonium nitrate (NH4NO3) [94], indicating that N status is important for OsphyB-mediated regulation of leaf senescence under light conditions.
N is one of the key macronutrients essential for plant growth and development [100]. The effects of light on N acquisition and use have been widely studied (summarized in [101]). In several plant species, including Arabidopsis, maize, soybean (Glycine max L.), sunflower (Helianthus annuus L.), tobacco (Nicotiana benthamiana), and wheat (Triticum aestivum L.), plant roots showed increased uptake of 15N-labeled nitrate (15NO3−) and nitrite (15NO2−) upon exposure to light [102][103]. Additionally, exogenous application of sugar to plant roots greatly increased NO3− and ammonium (NH4+) uptake [103][104]. These data suggest that the positive effects of light on the uptake of nutrients, including N compounds, are linked in part to the increase in carbohydrate supply from leaves to roots that result from the increase in photosynthesis caused by the increase in light availability. On the other hand, several reports implied that light signaling promotes the uptake of N as well as other nutrients. In Arabidopsis seedlings, the expression of ammonium transporter genes (AMT1;1, AMT1;2, and AMT2;1) was enhanced upon irradiation with red light [105]. HY5, a key transcription factor that positively regulates red-light signaling downstream of phyB was shown to bind to and activate the promoter of NITRATE TRANSPORTER2.1 (NRT2.1), encoding a high-affinity nitrate transporter [106]. Furthermore, genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) analysis indicated that HY5 directly binds to the promoters of genes associated with the uptake and assimilation of N and other nutrients [107]. Another ChIP-seq analysis showed that PIF4 also directly binds to the promoters of genes associated with nutrient uptake and assimilation, such as NRT1.1 and NRT1.2 as well as NIA2, which encodes a nitrate reductase [108]. Moreover, nitrate reductase activity was significantly higher in pif4 mutants than in the wild type, especially under red light [109], indicating that HY5 and PIF4 TFs positively or negatively regulate N uptake and assimilation under the regulation of phyB-mediated red-light signaling. It was recently shown that phyB/PIF4/PIF5/HY5-mediated red-light signaling is also involved in the promotion of phosphate (PO43−) uptake in roots by increasing the expression of phosphate transporter genes, including PHOSPHATE TRANSPORTER1;1 (PHT1;1) [110]. Thus, phyB-mediated red-light signaling promotes the uptake and utilization of some nutrients, including N and phosphorus (P), which negatively affects the initiation of leaf senescence.
3.5. Far-Red-Light Signaling-Mediated Regulation of Leaf Senescence
In contrast to red light, far-red light accelerates leaf senescence. Under shade (low red: far-red ratio), leaves exhibit accelerated leaf yellowing [111]. In addition, far-red light illumination decreases chlorophyll pigments and/or promotes leaf yellowing in various plant species, including liverwort (Marchantia polymorpha), cucumber, rice, and Pak choi (Brassica rapa ssp. chinensis) [77][112][113][114].
Far-red-light illumination diminishes red-light signaling by changing the active Pfr form to the inactive Pr form [69], which may, at least partially, contribute to the promotion of leaf yellowing. Indeed, the intermittent red-light pulse treatment strongly inhibited the increase in PIF4 and PIF5 protein levels in darkness, while pulses of red light followed by far-red light diminished this inhibitory effect [79]. In Pak choi leaves, far-red-light illumination enhanced, whereas red-light illumination reduced the expression of key SAGs, including EIN3 [114].
Lim et al. (2019) showed that under far-red light-enriched conditions, leaves of the Arabidopsis phyA loss-of-function mutant (phyA-211) turned yellow much faster than those of wild-type plant, while the phyB loss-of-function mutant (phyB-9) exhibited delayed leaf yellowing [115]. Under far-red light illumination, the expression of WRKY6 [116], was upregulated in phyA loss-of-function mutant but downregulated in phyB loss-of-function mutant [115]. Thus, phyA and phyB are involved in fine-tuning the far-red light-mediated promotion of leaf senescence by antagonistically regulating the expression of far-red light-inducible SAGs including WRKY6 (Figure 2).
FAR-RED IMPAIRED RESPONSE 1 (FAR1) and its closest homolog FAR-RED ELONGATED HYPOCOTYL3 (FHY3), which are transposase-derived TFs, play key roles in phyA-mediated far-red-light signaling pathways [117][118]. It was recently shown that FHY3 acts as a negative regulator of age-dependent and light signaling-mediated leaf senescence by directly repressing the transcription of WRKY28, which encodes a senescence-promoting WRKY TF [119]. Both fhy3 knockout mutant and WRKY28 overexpression lines exhibited early leaf yellowing under light conditions with high red: far-red ratio [119], suggesting that the FHY3–WRKY28 regulatory module plays an important role in the inhibition of leaf senescence under high red: far-red ratio.
3.6. Blue-Light Signaling-Mediated Regulation of Leaf Senescence
The effects of blue light on the initiation of leaf senescence appear to be much smaller than those of red light in some plant species. Postharvest senescence of broccoli (Brassica oleracea L.) was delayed by red-light-emitting diodes (LEDs) but was not affected by blue LEDs [120]. Similarly, red LEDs inhibited leaf yellowing in Pak choi, while the inhibitory effect of blue LEDs was much smaller [114].
The role of blue-light signaling on the initiation of leaf senescence is not yet well understood. CRYPTOCHROME 1 (CRY1) and (CRY2) are blue light photoreceptors that regulate a variety of blue-light-mediated biological processes [67]. The cry1 cry2 double mutant of Arabidopsis turned yellow, similar to the wild type, during dark-induced leaf senescence [79], indicating that cryptochrome-mediated blue light signaling plays little or role in dark-induced leaf senescence in Arabidopsis. On the other hand, in soybean, CRY2a acts as a negative regulator of leaf senescence; transgenic plants overexpressing CRY2a exhibited delayed leaf yellowing, whereas CRY2a RNAi plants exhibited accelerated-leaf-yellowing phenotype during the senescence phase [121]. In addition, CIB1 acts as an enhancer of leaf senescence [121]. Under blue light, CRY2a undergoes light-specific interaction with CIB1, which decreases the WRKY53b promoter-binding affinity of CIB1 [121]. Thus, CRY2a- and CIB1-mediated blue light signaling inhibits the initiation of leaf senescence in soybean. This shows that the effects of cryptochrome-mediated blue-light signaling on the initiation of leaf senescence varies among plant species.
In Arabidopsis, CRY1 directly interacts with PIF4 in a blue-light-dependent manner to repress its gene transcription in high-temperature-mediated hypocotyl elongation [122]. Thus, it is possible that CRY1-mediated blue-light signaling affects the initiation of leaf senescence under specific environmental stresses, such as high temperature, through the regulation of PIF4 activity, although this effect may be small compared with the effect of red light.
3.7. Connection between Circadian Rhythm and Light Signaling during Leaf Senescence
Recent studies show that circadian rhythm plays an important role in the regulation of leaf senescence. In Arabidopsis, the evening complex (EC), composed of ELF3, ELF4, and LUX APRHYTHMO (LUX), negatively regulates jasmonate (JA)-induced leaf senescence [123]. This EC was shown to directly bind to the promoter of MYC2, which encodes a key JA response regulator [124], and represses its expression, leading to the increase of expression of a number of JA-inducible genes [123]. In addition, plants carrying mutations in ELF3, ELF4, and LUX exhibited accelerated leaf yellowing during natural senescence and dark-induced leaf senescence [125] (Figure 2). In a diurnal rhythm, EC represses the expression of PIF4 and PIF5 early during the subjective night [126]. This EC–PIF4/PIF5 regulatory cascade is also important in dark-induced leaf senescence; the expression levels of PIF4 and PIF5 were upregulated in elf3 mutants but downregulated in ELF3 overexpressors after a few days of dark incubation [79].
It has recently been reported that CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), one of the core components of the circadian oscillator, negatively regulates leaf senescence by directly repressing the expression of ORE1, which acts downstream of PIF4 and PIF5 in the red-light signaling-mediated leaf senescence pathway [79], while directly activating the expression of GLKs [127] (Figure 2). PSEUDO-RESPONSE REGULATOR 9 (PRR9), which is another component of the circadian oscillator, promotes leaf senescence by directly enhancing ORE1 expression [125]. Thus, several components of the circadian oscillator, including ELF3, ELF4, LUX, CCA1, and PRR9 affect the expression of genes associated with red-light signaling-mediated leaf senescence, indicating that the circadian rhythm is closely linked with the transcriptional regulatory network of light signaling-mediated leaf senescence.
References
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf Senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136.
- Hörtensteiner, S. Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 2009, 19, 155–162.
- Guo, Y. Towards systems biological understanding of leaf senescence. Plant Mol. Biol. 2013, 82, 519–528.
- Sakuraba, Y.; Park, S.Y.; Paek, N.C. The divergent roles of STAYGREEN (SGR) homologs in chlorophyll degradation. Mol. Cells 2015, 38, 390–395.
- Woo, H.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf senescence: Systems and dynamics aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376.
- Kusaba, M.; Tanaka, A.; Tanaka, R. Stay-green plants: What do they tell us about the molecular mechanism of leaf senescence. Photisynth. Res. 2013, 117, 221–234.
- Quirino, B.F.; Normanly, J.; Amasino, R.M. Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol. Biol. 1999, 40, 267–278.
- Gepstein, S.; Glick, B.R. Strategies to ameliorate abiotic stress-induced plant senescence. Plant Mol. Biol. 2013, 82, 623–633.
- Zhang, H.J.; Dong, H.Z.; Li, W.J.; Zhang, D.M. Effects of soil salinity and plant density on yield and leaf senescence of field-grown cotton. J. Agric. Crop Sci. 2012, 198, 27–37.
- Jespersen, D.; Zhang, J.; Huang, B. Chlorophyll loss associated with heat-induced senescence in bentgrass. Plant Sci. 2016, 249, 1–12.
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56.
- Luoni, S.B.; Astigueta, F.H.; Nicosla, S.; Moschen, S.; Fernandez, P.; Heinz, R. Transcription factors associated with leaf senescence in crops. Plants 2019, 8, 411.
- Folta, K.M.; Childers, K.S. Light as a growth regulator: Controlling plant biology with narrow-bandwidth solid-state lighting systems. HortScience 2008, 43, 1957–1964.
- Kami, C.; Lorrain, S.; Hornistschek, P.; Fankhauser, C. Chapter two-light-regulated plant growth and development. Curr. Topics Dev. Biol. 2010, 91, 29–66.
- Makino, A.; Sakuma, H.; Sudo, E.; Mae, T. Differences between maize and rice in N-use efficiency for photosynthesis and protein allocation. Plant Cell Physiol. 2003, 44, 952–956.
- Hensel, L.L.; Grbić, V.; Baumgarten, D.A.; Bleecker, A.B. Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in arabidopsis. Plant Cell 1993, 5, 553–564.
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative, stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399.
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396.
- Waszczak, C.; Carmody, M.; Kanfasjärvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236.
- Dhindsa, R.S.; Dhindsa, P.P.; Thorpe, T.A. Leaf senescence: Correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 1981, 126, 93–101.
- Becerril, J.M.; Duke, S.O. Protopophyrin IX content correlates with activity of photobleaching herbicides. Plant Physiol. 1989, 90, 1175–1181.
- op dem Camp, R.G.L.; Przybyla, D.; Ochsenbein, C.; Laloi, C.; Kim, C.; Danon, A.; Wagner, D.; Hideg, E.; Göbel, C.; Feussner, I.; et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 2003, 15, 2320–2332.
- Piao, W.; Han, S.H.; Sakuraba, Y.; Paek, N.C. Rice 7-hydroxymethyl chlorophyll a reductase is involved in the promotion of chlorophyll degradation and modulates cell death signaling. Mol. Cells 2017, 40, 773–786.
- Pruzinská, A.; Tanner, G.; Anders, I.; Roca, M.; Hörtensteiner, S. Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron--sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 2003, 100, 15259–15264.
- Pruzinská, A.; Anders, I.; Aubry, S.; Schenk, N.; Taprtnoux-Lüthi, E.; Müller, T.; Kräutler, B.; Hörtensteiner, S. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 2007, 19, 369–387.
- Zimmermann, P.; Zentgraf, U. The correlation between oxidative stress and leaf senescence during plant development. Cell Mol. Biol. Lett. 2005, 10, 515–534.
- Jajic, I.; Sarna, T.; Strzalka, K. Senescence, stress, and reactive oxygen species. Plants 2015, 4, 393–411.
- Quan, L.J.; Zhang, B.; Shi, W.W.; Li, H.Y. Hydrogen peroxide in plants: A versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 2008, 50, 2–18.
- Deskain, R.; Nelli, S.J.; Hancock, J.T. Hydrogen peroxide-induced gene expression in Arabidopsis thaiana. Free Radic. Biol. Med. 2000, 28, 773–778.
- Balazadeh, S.; Wu, A.; Mueller-Roeber, B. Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal Behav. 2010, 5, 733–735.
- Garapati, P.; Xue, G.P.; Munné-Bosch, S.; Balazadeh, S. Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 2015, 168, 1122–1139.
- Kim, Y.S.; Sakuraba, Y.; Han, S.H.; Yoo, S.C.; Paek, N.C. Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol. 2013, 54, 1660–1672.
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612.
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.I.; Asensi-Fabado, M.A.; Munné-Bosch, S.; Antonio, C.; Tohge, T.; et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506.
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057.
- Ng, S.; Ivanova, A.; Duncan, O.; Law, S.R.; Van Aken, O.; De Clercq, I.; Wang, Y.; Carrie, C.; Xu, L.; Kmiec, B.; et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 2013, 25, 3450–3471.
- De Clercq, I.; Vermeirssen, V.; Van Aken, O.; Vandepoele, K.; Murcha, M.W.; Law, S.R.; Inzé, A.; Ng, S.; Ivanova, A.; Rombaut, D.; et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 2013, 25, 3472–3490.
- Van Aken, O.; De Clercq, I.; Ivanova, A.; Law, S.R.; Van Breusegem, F.; Millar, A.H.; Whelan, J. Mitochondrial and chloroplast stress responses are modulated in distinct touch and chemical inhibition phases. Plant Physiol. 2016, 171, 2150–2165.
- Lee, S.; Seo, P.J.; Lee, H.J.; Park, C.M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012, 70, 831–844.
- Sakuraba, Y.; Kim, D.; Han, S.H.; Kim, S.H.; Piao, W.; Yanagisawa, S.; An, G.; Paek, N.C. Multilayered regulation of membrane-bound ONAC054 is essential for abscisic acid-induced leaf senescence in rice. Plant Cell 2020, 32, 630–649.
- Robatzek, S.; Somissich, I.E. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001, 28, 123–133.
- Zhou, X.; Jiang, Y.; Yu, D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 2011, 31, 303–313.
- Scarpeci, T.E.; Zanor, M.I.; Carrillo, N.; Mueller-Roeber, B.; Valle, E.M. Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: A focus on rapidly induced genes. Plant Mol. Biol. 2008, 66, 361–378.
- Takahashi, M.A.; Asada, K. Superoxide anion permeability of phospholipid membranes and chloroplast thylakoids. Arch. Biochem. Biophys. 1983, 226, 558–566.
- Krasnovsky, A.A., Jr. Singlet molecular oxygen in photobiochemical systems: IR phosphorescence studies. Membr. Cell Biol. 1998, 12, 665–690.
- Triantaphylidès, C.; Krischke, M.; Hoeberichts, F.A.; Ksas, B.; Gresser, G.; Havaux, M.; Breusegem, F.V.; Mueller, M.J. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 2008, 148, 960–968.
- Gosti, F.; Beaudoin, N.; Serizet, C.; Webb, A.A.R.; Vartanian, N.; Giraudat, J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 1999, 11, 1897–1910.
- Babula, D.; Misztal, L.H.; Jakubowicz, M.; Kaczmarek, M.; Nowak, W.; Sadowski, J. Genes involved in biosynthesis and signalisation of ethylene in Brassica oleracea and Arabidopsis thaliana: Identification and genome comparative mapping of specific gene homologues. Theor. Appl. Genet. 2006, 112, 410–420.
- Weaver, L.M.; Gan, S.; Quirino, B.; Amasino, R.M. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 1998, 37, 455–469.
- Tanaka, A.; Ito, H.; Tanaka, R.; Tanaka, N.K.; Yoshida, K.; Okada, K. Chlorphyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc. Natl. Acad. Sci. USA 1998, 95, 12719–12723.
- Nakagawara, E.; Sakuraba, Y.; Yamasato, A.; Tanaka, R.; Tanaka, A. Clp protease controls chlorophyll b synthesis by regulating the level of chlorophyllide a oxygenase. Plant J. 2007, 49, 800–809.
- Sakuraba, Y.; Balazadeh, S.; Tanaka, R.; Mueller-Roeber, B.; Tanaka, A. Overproduction of chl B retards senescence through transcriptional reprogramming in Arabidopsis. Plant Cell Physiol. 2012, 53, 505–517.
- Sakuraba, Y.; Yokono, M.; Akimoto, S.; Tanaka, R.; Tanaka, A. Deregulated chlorophyll b synthesis reduces the energy transfer rate between photosynthetic pigments and induces photodamage in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 1055–1065.
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 2007, 19, 1362–1375.
- Horie, Y.; Ito, H.; Kusaba, M.; Tanaka, R.; Tanaka, A. Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J. Biol. Chem. 2009, 284, 17449–17456.
- Yamatani, H.; Kohzuma, K.; Nakano, M.; Takami, T.; Kato, Y.; Hayashi, Y.; Okumoto, Y.; Abe, T.; Kumamaru, T.; Tanaka, A.; et al. Impairment of Lhca4, a subunit of LHCI, causes high accumulation of chlorophyll and the stay-green phenotype in rice. J. Exp. Bot. 2018, 69, 1027–1035.
- Strand, A.; Asami, T.; Alonso, J.; Ecker, J.R.; Chory, J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 2003, 421, 79–83.
- Kindgren, P.; Norén, L.; López, J.D.; Shaikhali, J.; Strand, A. Interplay between Heat Shock Protein 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol. Plant 2012, 5, 901–913.
- Sakuraba, Y.; Schelbert, S.; Park, S.Y.; Han, S.H.; Lee, B.D.; Andrès, C.B.; Kessler, F.; Hörtensteiner, S.; Paek, N.C. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 2012, 24, 507–518.
- Schelbert, S.; Aubry, S.; Burla, B.; Agne, B.; Kessler, F.; Krupinska, K.; Hörtensteiner, S. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 2009, 21, 767–785.
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magunesium-dechelatase. Plant Cell 2016, 28, 2147–2160.
- Ono, K.; Kimura, M.; Matsuura, H.; Tanaka, A.; Ito, H. Jasmonate production through chlorophyll a degradation by Stay-Green in Arabidopsis thaliana. J. Plant Physiol. 2019, 238, 53–62.
- Ramel, F.; Birtic, S.; Ginies, C.; Soubigou-Taconnat, L.; Triantaphylidès, C.; Havaux, M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5535–5540.
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015, 84, 1114–1123.
- D’Alessandro, S.; Ksas, B.; Havaux, M. Decoding β-cyclocitral-mediated retrograde signaling reveals the role of a detoxification response in plant tolerance to photooxidative stress. Plant Cell 2018, 30, 2495–2511.
- McCREE, K.J. The action spectrum, absorptance and quantum yield of photosynthesis in crop plants. Agric. Meteorol. 1972, 9, 191–216.
- Lin, C. Plant blue-light receptors. Trends Plant Sci. 2000, 5, 337–342.
- Liang, T.; Yang, Y.; Liu, H. Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol. 2019, 221, 1247–1252.
- Li, J.; Li, G.; Wang, H.; Deng, X.W. Phytochrome signaling mechanisms. Arabidopsis Book 2011, 9, e0148.
- Sharrock, R.A.; Clack, T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 2002, 130, 442–456.
- Reed, J.W.; Nagatani, A.; Elich, T.D.; Fagan, M.; Chory, J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994, 104, 1139–1149.
- Leivar, P.; Quali, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28.
- Liu, H.; Liu, B.; Zhao, C.; Pepper, M.; Lin, C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011, 16, 684–691.
- Osterlund, M.T.; Hardtke, C.S.; Wei, N.; Deng, X.W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000, 405, 462–466.
- Biswal, U.C.; Sharma, R. Phytochrome regulation of senescence in detached barley leaves. Z. Phlanzenphysiol. 1976, 80, 71–73.
- Pfeiffer, H.; Kleudgen, H.K. Investigations on the phytochrome control of senescence in the photosynthetic apparatus of Hordeum Vulgae L. Z. Phlanzenphysiol. 1980, 100, 437–445.
- Tucker, D.J. Phytochrome regulation of leaf senescence in cucumber and tomato. Plant Sci. Lett. 1981, 23, 103–108.
- Biswal, U.C.; Kasemir, H.; Mohr, H. Phytochrome control of degreening of attached colyledons and primary leaves of mustard (Sinapis alba L.) seedlings. Photochem. Photobiol. 1982, 35, 237–241.
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636.
- Grbić, V.; Bleecker, A.B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995, 8, 595–602.
- Ray, S.; Mondal, W.A.; Choudhuri, M.A. Regulation of leaf senescence, grain-filling and yield of rice by kinetin and abscisic acid. Physiol. Plant. 1983, 59, 343–346.
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714.
- Bensmihen, S.; Rippa, S.; Lambert, G.; Jublot, D.; Pautot, V.; Granier, F.; Giraudat, J.; Parcy, F. The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 2002, 14, 1391–1403.
- Song, Y.; Yang, C.; Gao, S.; Zhang, W.; Li, L.; Kuai, B. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787.
- Zhang, Y.; Liu, Z.; Chen, Y.; He, J.X.; Bi, Y. PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Sci. 2015, 237, 57–68.
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399.
- Sakuraba, Y.; Kim, E.Y.; Han, S.H.; Piao, W.; An, G.; Todaka, D.; Yamaguchi-Shinozaki, K.; Paek, N.C. Rice phytochrome-interacting factor-like1 (OsPIL1) is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops. J. Exp. Bot. 2017, 68, 4103–4114.
- Wagner, D. Flower morphogenesis: Timing is key. Dev. Cell 2009, 16, 621–622.
- Mangan, S.; Alon, U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 2003, 100, 11980–11985.
- Waters, M.T.; Wang, P.; Korkaric, M.; Capper, R.G.; Saunders, N.J.; Langdale, J.A. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 2009, 21, 1109–1128.
- Shahnejat-Bushehri, S.; Tarkowska, D.; Sakuraba, Y.; Balazadeh, S. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2016, 2, 16013.
- Sakuraba, Y.; BülBül, S.; Piao, W.; Choi, G.; Paek, N.C. Arabidopsis EARLY FLOWERING3 increases salt tolerance by suppressing salt stress response pathways. Plant J. 2017, 92, 1106–1120.
- Kim, C.; Kim, S.J.; Jeong, J.; Park, E.; Oh, E.; Park, Y.I.; Lim, P.O.; Choi, G. High ambient temperature accelerates leaf senescence via PHYTOCHROME-INTERACTING FACTOR 4 and 5 in Arabidopsis. Mol. Cells 2020, 43, 645–661.
- Piao, W.; Kim, E.Y.; Han, S.H.; Sakuraba, Y.; Paek, N.C. Rice phytochrome B (OsphyB) negatively regulates dark- and starvation-induced leaf senescence. Plants 2015, 4, 644–663.
- Nakamura, Y.; Kato, T.; Yamashino, T.; Murakami, M.; Mizuno, T. Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa. Biosci. Biotechnol. Biochem. 2007, 71, 1183–1191.
- Sakuraba, Y.; Kim, E.Y.; Paek, N.C. Roles of rice PHYTOCHROME-INTERACTING FACTOR-LIE1 (OsPIL1) in leaf senescence. Plant Signal. Bahav. 2017, 12, e1362522.
- Sakuraba, Y.; Han, S.H.; Yang, H.J.; Piao, W.; Paek, N.C. Mutation of rice Early Flowering3.1 (OsELF3.1) delays leaf senescence in rice. Plant Mol. Biol. 2016, 92, 223–234.
- Rosado, D.; Trench, B.; Bianchetti, R.; Zuccarelli, R.; Rodrigues Alves, F.R.; Purgatto, E.; Segal Floh, E.I.; Silveira Nogueira, F.T.; Freschi, L.; Rossi, M. Downregulation of PHYTOCHROME-INTERACTING FACTOR 4 influences plant development and fruit production. Plant Physiol. 2019, 181, 1360–1370.
- Shi, Q.; Zhang, H.; Song, X.; Jiang, Y.; Liang, R.; Li, G. Functional characterization of the Maize phytochrome-interacting factors PIF4 and PIF5. Front. Plant Sci. 2018, 8, 2273.
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182.
- Sakuraba, Y.; Yanagisawa, S. Light signalling-induced regulation of nutrient acquisition and utilisation in plants. Semin. Cell Dev. Biol. 2018, 83, 123–132.
- Reed, A.J.; Canvin, D.T.; Sherrard, J.H.; Hageman, R.H. Assimilation of [15N]nitrate and [15N]nitrite in leaves of five plant species under light and dark conditions. Plant Physiol. 1983, 71, 291–294.
- Lejay, L.; Gansel, X.; Cerezo, M.; Tillard, P.; Müller, C.; Krapp, A.; von Wirén, N.; Daniel-Vedele, F.; Gojon, A. Regulation of root ion transporters by photosynthesis: Functional importance and relation with hexokinase. Plant Cell 2003, 15, 2218–2232.
- Lejay, L.; Tillard, P.; Lepetit, M.; Olive, F.D.; Filleur, S.; Daniel-Vedele, F.; Gojon, A. Molecular and functional regulation of two NO3- uptake systems by N- and C-status of Arabidopsis plants. Plant J. 1999, 18, 509–519.
- Huang, L.; Zhang, H.; Zhang, H.; Deng, X.W.; Wei, N. HY5 regulates nitrite reductase 1 (NIR1) and ammonium transporter1:2 (AMT1;2) in Arabidopsis seedlings. Plant Sci. 2015, 238, 330–339.
- Chen, X.; Yao, Q.; Gao, X.; Juang, C.; Harberd, N.P.; Fu, X. Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 2016, 26, 640–646.
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749.
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809.
- Jonassen, E.M.; Sandsmark, B.A.A.; Lillo, C. Unique status of NIA2 in nitrate assimilation: NIA2 expression is promoted by HY5/HYH and inhibited by PIF4. Plant Signal. Bahav. 2009, 4, 1084–1086.
- Sakuraba, Y.; Kanno, S.; Mabuchi, A.; Monda, K.; Iba, K.; Yanagisawa, S. A phytochrome-B-mediated regulatory mechanism of phosphorus acquisition. Nat. Plants 2018, 4, 1089–1101.
- Brouwer, B.; Ziolkowska, A.; Bagard, M.; Keech, O.; Gardeström, P. The impact of light intensity on shade-induced leaf senescence. Plant Cell Environ. 2012, 35, 1084–1098.
- De Greef, J.A.; Fredericq, H. Enhancement of senescence by far-red light. Planta 1972, 104, 272–274.
- Okada, K.; Katoh, S. Two long-term effects of light that control the stability of proteins related to photosynthesis during senescence of rice leaves. Plant Cell Physiol. 1998, 39, 394–404.
- Song, Y.; Qiu, K.; Gao, J.; Kuai, B. Molecular and physiological analyses of the effects of red and blue LED light irradiation on postharvest senescence of pak choi. Postharvest Biol. Technol. 2020, 164, 111155.
- Lim, J.; Park, J.H.; Jung, S.; Hwang, D.; Nam, H.G.; Hong, S. Antagonistic roles of phyA and phyB in far-red light-dependent leaf senescence in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1753–1764.
- Robatzek, S.; Somssich, I.E. Targets of AtWKRY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002, 16, 1139–1149.
- Wang, H.; Deng, X.W. Arabidopsis FHY3 defines a key phytochrome A signaling components directly interacting with its homologous partner FAR1. EMBO J. 2002, 21, 1339–1349.
- Wang, H.; Wang, H. Multifaceted roles of FHY3 and FAR1 in light signaling and beyond. Trends Plant Sci. 2015, 20, 453–461.
- Tian, T.; Ma, L.; Liu, Y.; Xu, D.; Chen, Q.; Li, G. Arabidopsis FAR-RED ELONGATED HYPOCOTYL3 integrates age and light signals to negatively regulate leaf senescence. Plant Cell 2020, 32, 1574–1588.
- Ma, G.; Zhang, L.; Setiawan, C.K.; Yamawaki, K.; Asai, T.; Nishikawa, F.; Maezawa, S.; Sato, H.; Kanemitsu, N.; Kato, M. Effect of red and blue LED light irradiation on ascorbate content and expression of genes related to ascorbate metabolism in postharvest broccoli. Postharvest Biol. Technol. 2014, 94, 97–103.
- Meng, Y.; Li, H.; Wang, Q.; Liu, B.; Lin, C. Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell 2013, 25, 4405–4420.
- Ma, D.; Li, X.; Guo, Y.; Chu, J.; Fang, S.; Yan, C.; Noel, J.P.; Liu, H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 2016, 113, 224–229.
- Zhang, Y.; Wang, Y.; Wei, H.; Li, N.; Tian, W.; Chong, K.; Wang, L. Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol. Plant 2018, 11, 326–337.
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703.
- Kim, H.; Kim, H.J.; Vu, Q.T.; Jung, S.; McClung, C.R.; Hong, S.; Nam, H.G. Circadian control of ORE1 by PRR9 positively leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 8448–8453.
- Nusinow, D.A.; Helfer, A.; Hamilton, E.E.; King, J.J.; Imaizumi, T.; Schultz, T.F.; Farré, E.M.; Kay, S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control hypocotyl growth. Nature 2011, 475, 398–402.
- Song, Y.; Jiang, Y.; Kuai, B.; Li, L. CIRCADIAN CLOCK-ASSOCIATED 1 inhibits leaf senescence in Arabidopsis. Front. Plant Sci. 2018, 9, 280.




