| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Boris Andryukov | + 1573 word(s) | 1573 | 2021-01-21 04:28:37 | | | |
| 2 | Bruce Ren | + 274 word(s) | 1847 | 2021-01-28 02:37:16 | | |
Video Upload Options
The coronavirus (covid-19) pandemic in 2019 caused by sars-cov-2 highly infectious virus has triggered the global health and economic crisis. Controlling the spread of disease requires an effective and extensive laboratory strategy to test the population through multiple platforms to ensure rapid and accurate testing. diagnosis.
1. Introduction
By the end of 2019, a new virus named cowid-19 has appeared in China for the first time. 1.2.[1][2] Subsequently, globally, the epidemic has become a catastrophic pandemic and caused serious crises in the world health and economic system. The virus spreads easily from person to person through the air, so it spreads rapidly in densely populated areas. According to the data of the European Center for Disease Control and prevention, in November 2020, there will be more than 62 million cases of cowid-19 worldwide, of which 14 million will die [2][3][4]
Similar to the clinical manifestations of other ozone depleting substances, the possibility of asymptomatic diseases, Due to the relatively high infection rate, it is difficult to carry out epidemiological monitoring of the spread of the virus. In this regard, the platform for the effective diagnosis of covid-19 is of great significance. Because they ensure timely detection and treatment of patients and monitoring of epidemics, [3] 4. Use the experience of other recent viral epidemics.
Three new pathogenic coronaviruses have emerged in the 21st century, which is of great concern. This is coronaviridae, including mers CoV, previously described pathogen of Middle East respiratory syndrome) mers, Jordan, 2012, and sars-cov-1, pathogen of acute respiratory syndrome. SARS in China, 2002) [5][6][7] in addition, some α - strains (α and β - Coronavirus) [8].
A genome-wide sequencing and phylogenetic analysis showed that the causative agent of COVID-19 is a β-coronavirus of the same subgenus as the SARS-CoV that has a rounded shape with a diameter of 60 to 140 nm [9]. To successfully control a pandemic, in addition to studying the viral agent, it is necessary to identify the main mechanisms of infection and determine the key strategies for diagnosing the infection.
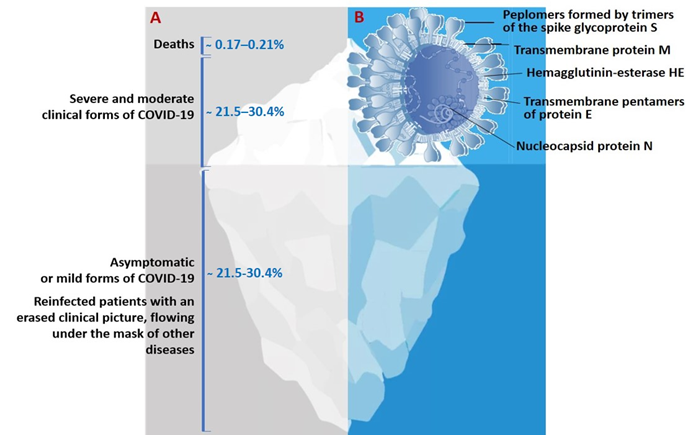
The genome of SARS-CoV-2 is a single-stranded positive-sense RNA of 29,903 nucleotides [10][11]. This genome encodes as many as 27 structural and non-structural proteins that provide transcription and replication of the virus (genes ORFlab and ORFla), as well as its pathogenic effects. The viral proteome includes polyproteins, structural and non-structural proteins. Some of the structural proteins such as, primarily, the spike glycoprotein (S) exposed on the phospholipid membrane, as well as the envelope protein (E), membrane protein (M), and nucleocapsid (N), are of particular biotechnological, pharmacological, and biomedical interest [12] (Figure 1B).
Figure 1. (A)The actual picture of the spread of COVID-19, resembling an iceberg and (B) some of the SARS-CoV-2 structural proteins of biotechnological and pharmacological interest.
The results of a molecular phylogenetic analysis showed that the genomes of SARS-CoV-2 and SARS-CoV, are related with an approximately 80% similarity. In particular, they share the largest of the structural proteins, glycoprotein S, which protrudes from the surface of mature virions. The S-protein plays key roles in virus attachment, fusion, and entry into human cells. For this purpose, the virus uses receptor-binding domain (RBD), which mediates binding to angiotensin converting enzyme 2 (ACE2) [13][14][15]. The highly immunogenic receptor-binding domain (RBD) of this protein is the main target for the neutralizing activity of antibodies and serves as a basis for the development of vaccines [16][17][18].
2. Laboratory Testing as A Basis for the Diagnosis, Treatment, and Monitoring of COVID-19
One of the most important issues in the strategy to control the new infection has been the necessity of mass laboratory-based screening of populations exposed to high risk of infection. The timely and high-quality laboratory-based diagnostics of patients infected by SARS-CoV-2 has become the top priority in eliminating the pandemic and taking quarantine measures [16]. Under these conditions, the creation of fast, effective, and inexpensive diagnostic tools is a necessary part of the fight against the new infection. When diagnosing COVID-19, the major challenge that the healthcare system faces is to identify the role and place of various diagnostic platforms for screening, diagnosing, and monitoring new coronavirus infections [19].
The results of the SARS-CoV-2 genome sequencing have become a basis for the development of vaccines and test systems to provide diagnosis and epidemic monitoring of the infection [19]. However, with lack of experience in eliminating the COVID-19 pandemic, the healthcare system now faces new issues and problems such as timeliness, frequency, and choice of testing tools, as well as identification of the place and role of their results in decision-making. These questions can be answered through solving the issues of availability of certain types of laboratory tests, timeliness of testing and their informativeness, and also the clinical, epidemiological and economic feasibility of their use in the rapidly changing and unprecedented pattern of the spread of the pandemic in recent history [20][21][23].
To date, there are substantial differences in the choice of optimal diagnostic tools and effective methods for testing patients with COVID-19, their contacts, asymptomatic vectors of the virus, medical specialists and other representatives of emergency medical services [22]. After nearly a year of fighting COVID-19, healthcare efforts are still measured in terms of number of tests performed [22].
The dynamics and sensitivity of laboratory tests for this category of patient remains unstudied, and they can become a source of infection for others [23]. This suggests that laboratory-based diagnostic strategies aimed at patients with symptoms are not sufficient to prevent the spread of the virus [20][21][22].
Negative polymerase chain reaction (PCR) tests and the detection of presence of specific antibodies are considered the criteria signifying a recovered patient, without considering the consequences on their health and quality of life. Nevertheless, levels of antibodies in those who have recovered are not further investigated, and the intensity of immunity and the possibility of re-infection by COVID-19 also remain unstudied [21]. This is probably due to the lack of understanding of the immune signaling pathways triggered by SARS-CoV-2, as well as the general immunopathology of this infection [22].
Consequently, a rapid, complete, and most accurate assessment of the spread of the virus requires laboratory-based tests for total screening of the population, which will allow rapid identification and isolation of infected patients. In addition, there is a necessity for a long-term strategy for preventing recurrent outbreaks of infection, which would imply repeated and regular mass testing of the immune response in the population to determine the effectiveness of vaccination [24].
The situation regarding the diagnostics of COVID-19 is further complicated by the lack of awareness in society, the mass media, among medical officials and some biomedical specialists concerning the differences between the existing types of diagnostic tests for this infection. Therefore, it is not surprising that neither a unified methodology with clear goals and objectives, nor an agreed interpretation of the results obtained yet exists [25].
Obviously, one of the major issues in the development of a testing strategy during the COVID-19 pandemic is associated with the existing types of diagnostic tool and their fundamental difference, clinical practicability, and uselessness for certain categories of patients at different stages of the disease. Other widely discussed issues are timing of testing, frequency, and correct interpretation of results obtained.
3. Laboratory-Based Tests to Diagnose COVID-19
Data obtained from routine laboratory examinations are non-specific (leukopenia, lymphopenia, mild thrombocytopenia, increased levels of acute phase proteins, decreased partial pressure of oxygen in the blood, and, in severe cases, identification of markers of cytokine storm in the form of increased levels of cytokines IL2, IL4, IL6, IL7, IL10, and TNF-α). These tests are helpful in treatment of patients diagnosed as COVID-19 positive.
All currently existing types of special laboratory tests for diagnosing COVID-19 can be divided into two categories: those that directly detect the virus (its genome or antigens) and those that detect the human immune response to its presence (antibodies IgM, IgA, and IgG).
Laboratory-based tests for COVID-19 are used for a variety of purposes. A diagnostic examination is carried out for patients with clinical symptoms (complaints) in order to confirm the diagnosis. A screening study is carried out for people who feel healthy in order to identify disease among them (including the asymptomatic form of infection). At last, monitoring is carried out for patients undergoing treatment in order to assess the effectiveness or dynamics of the latter.
With the lack of specific symptoms and lack of proven effectiveness of etiotropic treatment and vaccination methods, the results of special laboratory diagnostics are the only source of data to confirm the presence and provide monitoring of the progress of COVID-19 [26].
The main analytical characteristics of laboratory-based tests are their sensitivity (which is evaluated as the probability of positive result in a patient with the disease) and specificity (negative test results in a healthy person). In addition, the effectiveness of tests is evaluated by their predictive value: the post-test probability of the disease in persons with a positive test result and its absence in persons with a negative test result.
Most test system manufacturers report high analytical performance (90%–100%) in cases in which their test systems are used under ideal conditions. However, in an actual situation, the diagnostic efficiency of a test depends on a number of factors (such as the clinical form of COVID-19, the duration of the disease, the quality of collection and the type of biomaterial, the conditions of its storage, transportation, etc.).
In an ideal (hypothetical) case, when using a test that detects SARS-CoV-2 and has 100% sensitivity and specificity, it would be possible to survey the entire world’s population. Depending on the results obtained, all infected patients can be sorted out and divided into the following categories: asymptomatic carriage and, depending on the clinical manifestation, mild, moderate and severe COVID-19. Accordingly, all patients with positive tests, depending on the clinical signs, are isolated either for quarantine, or home treatment, or treatment at a medical unit.
Alternatively, in another hypothetical case, the entire population is screened for the presence of IgG antibodies against SARS-CoV-2 using another test that has a 100% sensitivity and specificity to identify patients who were previously infected but had the asymptomatic form or were immune to the virus. These categories of the population, with their antibody level regularly monitored, could be recruited as volunteer to provide social or medical assistance to diseased people.
The actual pattern of distribution of COVID-19 resembles an iceberg, where the categories of seriously ill and hospitalized patients are in the smaller, above-water, part, and those who die from infection are at the very top (Figure 1). The largest proportion of the underwater part of the iceberg is represented by patients who have had an infection in an asymptomatic form of the disease which, depending on gender and age, account for more than 78%, or the mild form, without specific clinical manifestation, or with symptoms of acute respiratory or other flu-like infections [27 (Figure 1A). In this case, asymptomatic patients bear the same viral load for the same period of time as those with the pronounced form of infection and are considered as the main source of infection spread.
Although the above cases are hypothetical, they can help to determine the situation and evaluate the diagnostic value of available tests, In addition, especially when there is no necessary therapeutic agent or vaccine, the feasibility of using these vaccines is very high.
References
- World Health Organization (WHO) Official Website. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200701-covid-19-sitrep-163.pdf?sfvrsn=c202f05b2 (accessed on 2 September 2020).
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. doi:10.1038/s41564-020-0695-z.
- Centers for Disease Control and Prevention. 2019 Novel Coronavirus, Wuhan, China. Information for Healthcare Profes-sionals. Available online: https://www.cdc.gov/coronavirus/2019-nCoV/hcp/index.html (accessed on 11 September 2020).
- European Centre for Disease Prevention and Control (ECDC) COVID 19. Available online: https:// www.ecdc.europa.eu/en/novel-coronavirus-china (accessed on 11 August 2020).
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Charac-teristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. doi:10.1056/NEJMoa2002032.
- Dinnes, J.; Deeks, J.J.; Adriano, A.; Berhane, S.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; Beese, S.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020, 8, CD013705. doi:10.1002/14651858.CD013705.
- Wu, J.-L.; Tseng, W.-P.; Lin, C.-H.; Lee, T.-F.; Chung, M.-Y.; Huang, C.-H.; Chen, S.-Y.; Hsueh, P.-R.; Chen, S.-C. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020, 81, 435–442. doi:10.1016/j.jinf.2020.06.023.
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1. In Reference Module in Life Sciences; Elsevier: Oxford, UK, 2020. doi:10.1016/B978-0-12-809633-8.21501-X.
- Ghebreyesus, T.A.; Swaminathan, S. Scientists are sprinting to outpace the novel coronavirus. Lancet 2020, 395, 762–764.
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328.
- Calisher, C.; Carroll, D.; Colwell, R.; Corley, R.B.; Daszak, P.; Drosten, C.; Enjuanes, L.; Farrar, J.; Field, H.; Golding, J.; et al. Statement in support of the scientists, public health professionals, and medical professionals of China combatting COVID-19. Lancet 2020, 395, e42–e43. doi:10.1016/S0140-6736(20)30418-9.
- Fernandes, J.D.; Hinrichs, A.S.; Clawson, H.; Gonzalez, J.N.; Lee, B.T.; Nassar, L.R.; Raney, B.J.; Rosenbloom, K.R.; Nerli, S.; Rao, A.A.; et al. The UCSC SARS-CoV-2 Genome Browser. Nat. Genet. 2020, 52, 1–8. doi:10.1038/s41588-020-0700-8.
- Hoffman, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. doi:10.1016/j.cell.2020.02.052.
- Letko, M.; Marzi, A.; Munster, V.J. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. doi:10.1038/s41564-020-0688-y. ACE2
- Walls, A.C.; Tortorici, M.A.; Snijder, J.; Xiong, X.; Bosch, B.-J.; Rey, F.A.; Veesler, D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. USA 2017, 114, 11157–11162. doi:10.1073/pnas.1708727114.
- DeKosky, B.J. A molecular trap against COVID-19. Science 2020, 369, 1167–1168. doi:10.1126/science.abe0010.
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. doi:10.1038/s41586-020-2180-5.
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. doi:10.1016/j.cell.2020.08.012.
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection. Ann. Intern. Med. 2020, 173, 362–367. doi:10.7326/M20-3012.
- Petrosillo, N.; Viceconte, G.; Ergonul, O.; Ippolito, G.; Petersen, E. COVID-19, SARS and MERS: Are they closely related? Clin. Microbiol. Infect. 2020, 26, 729–734.
- Noh, J.Y.; Yoon, S.W.; Kim, D.J.; Lee, M.S.; Kim, J.H.; Na, W.; Song, D.; Jeong, D.G.; Kim, H.K. Simultaneous detection of severe acute respiratory syndrome, Middle East respiratory syndrome, and related bat coronaviruses by real-time reverse transcription PCR. Arch. Virol. 2017, 162, 1617–1623.
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020, 368, 489–493. doi:10.1126/science. abb3221.
- Li, N.; Wang, P.; Wang, X.; Geng, C.; Chen, J.; Gong, Y. Molecular diagnosis of COVID 19: Current situation and trend in China (Review). Exp. Ther. Med. 2020, 20, 1. doi:10.3892/etm.2020.9142.
- Yang Y, Yang M, Shen C, et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv; 2020. doi: 10.1101/2020.02.11.20021493.
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357.
- Chen, Z.; Zhang, Z.; Zhai, X.; Li, Y.; Lin, L.; Zhao, H.; Bian, L.; Li, P.; Yu, L.; Wu, Y.; et al. Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal Chem. 2020, 92, 7226–7231.
- Terpos, E.; Ntanasis‐Stathopoulos, I.; Elalamy, I.; Kastritis, E.; Sergentanis, T.N.; Politou, M.; Psaltopoulou, T.; Gerotziafas, G.; Dimopoulos, M.A. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020, 95, 834–847.