
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Kristina Malsagova | + 240 word(s) | 240 | 2020-12-28 09:12:36 | | | |
| 2 | Kristina Malsagova | + 3602 word(s) | 3842 | 2021-01-05 15:38:59 | | | | |
| 3 | Kristina Malsagova | + 3602 word(s) | 3842 | 2021-01-05 15:39:09 | | | | |
| 4 | Vivi Li | Meta information modification | 3842 | 2021-01-07 04:40:30 | | |
Video Upload Options
Pharmacogenomics is a study of how the genome background is associated with drug resistance and how therapy strategy can be modified for a certain person to achieve benefit. The pharmacogenomics (PGx) testing becomes of great opportunity for physicians to make the proper decision regarding each non-trivial patient that does not respond to therapy. Although pharmacogenomics has become of growing interest to the healthcare market during the past five to ten years the exact mechanisms linking the genetic polymorphisms and observable responses to drug therapy are not always clear. Therefore, the success of PGx testing depends on the physician’s ability to understand the obtained results in a standardized way for each particular patient. The review aims to lead the reader through the general conception of PGx and related issues of PGx testing efficiency, personal data security, and health safety at a current clinical level.
1. Introduction
P4-medicine represents an actively developing field of modern medical science. The P4 conception is based on a personalized approach to human health (Personalization, Prediction, Prevention, and Participation). Modern diagnostic kits allow the identification of human metabolic characteristics at the molecular level, thus enabling the revelation of a personal, genetically determined predisposition to a disease or certain metabolic disorders in particular individuals [1]. Personalized medicine involves drug therapy to improve the patient’s condition and minimize any adverse effects, thus increasing the quality of life at both the individual and socioeconomic levels.
Pharmacogenetics goes back to 1959 when Vogel coined this term to designate severe adverse drug reactions in a small number of patients reported by the pharmacologists [2]. The adverse reactions, which followed the administration of primaquine, succinylcholine, and isoniazid, were anemia, apnea, and peripheral neuropathy, respectively [3][4].
Pharmacogenetics is purposed to study the response to the drug therapy depending on the genetic background. Response to drugs is frequently governed by genes encoding drug-metabolizing proteins, thus, regulating drug transformation, pharmacokinetics, and pharmacodynamics [5].
PGx may support the investigation of the effect of vitamins [6][7] and additives/supplements and, to some degree [8], homeopathic preparations. However, there is no strong evidence regarding the interaction between genes and vitamins/supplements/homeopathies.
The majority of the related assays in PGx are erroneous or misinterpreted due to biases in the design of the experiment, small sampling, small size of the population, and insufficient time of observation. So far, it is obligatory to provide more correct experiments and researches to observe the suggested effects, otherwise, most of the discussion about the possible influence of non-pharmacological compounds on the genome could be considered doubtful.
In clinical practice, a physician follows the national standards of specialized medical care, based on the evidence from fundamental research and clinical trials of drugs. However, to a greater extent, the therapy process remains a creative task [9].
In addition to adverse drug reactions, the body can demonstrate immunity and/or just partial response to the treatment [10]. Despite the underlying causes of drug resistance remain unclear; however, one can suppose that this is connected with genetic factors. Moreover, aside from the predisposition to diseases, various body metabolic functions are also determined genetically. Namely, genetic variations probably determine the rates of synthesis and decay of multiple biomolecules in the body, the effect of pharmaceuticals, the metabolism of nutrients, etc. [11]. However, answers to these questions have not yet been received.
Nevertheless, genetic testing is slowly finding its niche in drug therapy selection—this process is followed by improving care to a widening range of patients. Pharmacogenomic Biomarkers in Drug Labeling Food and Drug Administration (FDA) provides data about 297 drugs for 100 molecular biomarkers (www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling). Several companies offer genetic testing for adverse drug reactions in patients.
2. Prospects for the Introduction of a Pharmacogenetic Test into the Clinic.
Nowadays, personalized medicine becomes more and more important [12]. Ongoing clinical trials can result in the introduction of pharmacogenetic testing into practice. This may accelerate the approval of distinct medication with no obligation to be tested on a PGx matter, which makes their market entry faster and more cost-effective. Typically, genomic information related to individual patients is made available to those who prescribe therapy. By 2017, The UK planned to perform the sequencing of 100,000 genomes of cancer patients and patients suffering from occasional or most dangerous infectious diseases (HIV, hepatitis C, tuberculosis), and to provide the pharmacogenetic information about the patients admitted in the study by the National Health Service [13]. Company 23andMe (Sunnyvale, CA, USA) (www.23andme.com) also provides pharmacogenetic information to guide the treatment directly upon customer request [14]. The FDA approved the first pharmacogenetic test in 2005. That was AmpliChip CYP450 test system manufactured by Affymetrix (Santa Clara, CA, USA) and Third Wave Technologies (Madison, WI, USA) Invader UGT1A1 (UDP-glucuronosyltransferase) Molecular Assay. The approval procedure was endorsed by clinicians, general healthcare systems, insurance companies, and concerned patients to determine the best way to integrate these tests into clinical practice [15]. At the same time, the critical questions raised were as to whom the tests should be applied and what are the most appropriate circumstances for their application, what evidence is required for the application of these tests, and how the results of the tests must be stored in electronic health data depositaries [16]? Haga and Kantor [17] reviewed laboratories, which offer clinical PGx testing in the United States. Of the 111 reviewed laboratories, 76 offered PGx testing services. Of these laboratories, 31 laboratories offered tests for only specific genes; 30 laboratories offered tests for multiple genes, while only 15 laboratories offered both types of tests. A total of 45 laboratories offered 114 multigene panel tests that cover 295 genes. However, no clinical guidelines were available for most of these tests [17]. In the industry, there is a trend towards multiplex tests intended to detect polymorphisms in a large number of genes. In 2005, the FDA-approved AmpliChip test (Roche, Basel, Switzerland) was designed to analyze two genes [18]. In 2010, Affymetrix introduced the DMET chip to diagnose 225 genes; the latter number is now expanded to 231 genes [17][19]. In 2012, researchers from Stanford University and the University of Florida developed a panel containing an SNP array of 120 genes including 25 genes responsible for drug metabolism and 12 drug carrier genes [20]. In 2014, the PGRN-Seq capture test for the analysis of 84 pharmacogens was developed [21]. The variety of gene panels is not limited to the examples mentioned above. Other types of pharmacological tests are available or are under development [22][23].
Currently, the clinical relevance of multigene panels mainly depends on a few well-studied and classical genes. Using the 84 gene PGRN-Seq capture panel, the examination of only five genes among ca. 5000 patients indicated that 99% of tested patients carried at least one clinically valid variant or one known variant relevant to decide about their treatment [24]. However, the clinical significance of large multiplex panels can mainly be determined by a certain task to be solved. For instance, PGx testing can be arranged as an immediate decision on treatment based on a panel of genes with a high level of evidence for a particular drug. However, the most common type of multigenic complex panel PGx test without specific clinical indications can be performed for a forward-looking patient. Such tests can be warranted since alternative drugs can be further used, and the clinical relevance of the data can increase with time. Despite the absence of consensus on the preventive PGx testing [25][26], many healthcare organizations implemented such testing programs to obtain valuable information regarding clinical validity and usefulness [21][27].
2.1. Audience for PGx Testing
Currently, PGx tests exist in various areas of medicine, including, but not limited to, psychiatry, cardiology, anesthesia, and oncology. Some clinical guidelines for PGx tests are accessible for the prediction of tricyclic antidepressants (TCAs) and selective serotonin reuptake inhibitor (SSRI) efficacy based on CYP2D6 and CYP2C19 activity [28][29]. Recent studies revealed reduced adverse effects and improved scores in depressed patients after PGx-based antidepressant therapy [30]. PGx testing is particularly attractive given the time frame required. For instance, the estimation of the complete therapeutic response to SSRIs can require 4 to 6 weeks [31]. The patient and the healthcare provider can spend several months adjusting the dose and/or prescribing new medications before it becomes clear that the therapy does not lead to the therapeutic effect. PGx testing can allow a physician to determine the best drug for a given situation in a much shorter time.
PGx testing applicability, to a certain extent, depends on the intensity of potential adverse reactions to the drug. For instance, Abacavir, used for HIV treatment, can produce severe cutaneous adverse reactions (SCAR) [32]. Generally, the risk is low, but HLA-B*57:01 variation is related to much more pronounced SCAR after Abacavir intake. Thus, this drug is contraindicated for HLA-B*57:01- positive patients [32].
The use of PGx testing is relevant to the selection of the Warfarin dose. Intake of dietary vitamin K, health and social conditions, and genetic variations were also found to affect Warfarin therapy [33]. Changes in CYP2C9 can disrupt the metabolism of Warfarin, and alterations in VKORC1 (vitamin K epoxide reductase) can increase the drug susceptibility of a patient [32][33].
The use of codeine was limited to adult patients after the evidence of a risk of increased adverse effects in pediatric patients. The study revealed adverse reactions in infants whose breastfeeding mothers underwent codeine therapy [34]. These reactions resulted from codeine conversion to morphine, performed mainly by CYP2D6 protein [35]. Similar reactions can occur with other CYP2D6-mediated pain relievers—such as tramadol, oxycodone, and hydrocodone [36].
The high interest of patients in PGx testing was revealed [37][38]. The patients are particularly interested in the possibility of using recommendations based on PGx test results to reduce adverse drug effects and to choose proper therapy [39]. However, the cost of the tests, the insurance coverage, and the availability of testing results represent the limitations for PGx testing [40][41]. Some questions are still relayed uncertain after PGx testing. Thus, patients should be appropriately informed about the capabilities of PGx testing [42].
For this reason, one should understand that PGx testing allows the identification of (1) drugs with an increased risk of causing adverse effects, (2) drugs with a narrow therapeutic index. Besides that, PGx testing can reduce the set of drugs for therapy and predict the drug dosage [42][43]. At the same time, PGx testing will not be efficient for predicting: (1) occurrence of all possible adverse reactions with a drug, (2) the risk of a specific adverse effect for all drugs, 3) the risk of occurrence of complications [43].
The PGx testing provides an opportunity in decision making of whether the chosen medication and treatment strategy is of advantage and gives the proper results over the expectations based on the obtained profile of patients. That is exactly what opens the door for personalized medicine, that is what should happen when a person has a choice: to be healed but not at a cost of health deterioration, not in awaiting while inappropriate drug boosts dire consequences instead of a satisfying outcome.
2.2. Choice of PGx Testing
PGx testing can be carried out either as a single gene analysis or as a multiplex panel of ten or more genes. Early testing mostly involves analyzing multiple variants of the same gene, targeting the most common and most effective variants. Novel technologies significantly increased the number of genes and variations covered by a single test. Most PGx tests analyze a variety of clinically relevant SNPs. During the determination of the best test or panel for a patient (or a population), one should consider the therapeutic indication. For instance, testing for CYP2D6 and CYP2C19 is required for antidepressant therapy. In addition, it will also be useful to consider whether the patient would benefit from a PGx test for cardiovascular and pain-killing drugs [44]. A panel test can be more expensive than a single gene assay, but it ensures that co-prescribed drugs are also tested.
Panel PGx tests are heterogeneous and vary in volume and scope [45][46]. Most of the panels cover several of the best-studied and most potent genes. The panel can contain SNP combinations based on a literature review of prospective studies. A panel that includes many genes may not necessarily provide additional value to the patient, as not all options offer the same clinical relevance. Some variants can be extremely rare outside of certain populations but can be quite common within a certain group. For instance, the HLA-B * 15: 02 variant, associated with an increased risk of SCAR in patients prescribed carbamazepine, has an allele frequency of 0.04% in patients of European descent and 6.88% in patients of East Asian descent [47]. A panel can be more beneficial to the patient if it analyzes options that match their ethnic origin more closely.
One should keep in mind available alternatives or special analyses. Multiple copies of the CYP2D6 gene (e.g., duplications) occur in about 1 in 8 patients, and this number may be even higher in black and Asian patients [48]. Gene duplication can cause increased enzymatic activity and can be clinically relevant. However, not all panels can reveal the presence or degree of gene duplication.
In addition to the panel contents, one should consider such factors as the type of biomaterial (buccal smear, saliva, or blood, etc.) since the way of biomaterial sampling can pose a problem for the patient. The access to results and the methods of their obtaining also represent the factors that should be considered. Several panels provide nothing but raw genetic data, while others are fully integrated into the electronic health record and enable sophisticated clinical decision support systems (CDS). Finally, one should consider the potential cost of PGx testing. Patients vary in ways and means to pay for testing.
2.3. Interpreting PGx Test Results
When prescribing PGx testing, one should keep in mind that the result obtained represents only a part of the overall picture of the patient’s condition. Therefore, to determine the therapy risks and benefits, the PGx test results should be used and interpreted by taking into account the state of all the patient’s systems, concomitant medications, and current pathological conditions. When the “best” drug is identified with the PGx test, this does not necessarily mean that it should be used in therapy since the patient may have a history of severe adverse reactions to the drug. Conversely, the PGx test identification of an increased risk of therapeutic failure should not lead to drug discontinuation if the current therapy is effective. Different result structures can be used, depending on the gene and protein in question [49]. Some genes can be described in terms of metabolic activity, some by their general function, and others only as present or absent. Several PGx test results describe general gene function—such as SLCO1B1 (solute carrier organic anion transporter family member 1B1), associated with simvastatin; VKORC1 (vitamin K epoxide reductase complex subunit 1), associated with Warfarin; OPRM1 (opioid receptor Mu 1), associated with opioids [49]. Results for these genes can be reported as normal, intermediate, or low function. For instance, a “normal gene function” result indicates that no change in the patient’s dosage regimen is required. In other cases (decreased or poor function), the physician’s recommendations will be based on the information on a reduced functional activity (or complete inactivity) of the analyzed genes.
PGx test results can be “positive” or “negative” [50]. For instance, human leukocyte antigen (HLA) genes produce essential components of the immune system. Patients who are positive for HLA-B * 58: 01, run an increased risk of hypersensitivity to allopurinol; patients, who are positive for HLA-B * 15: 02, run an increased risk of SCAR with carbamazepine or oxcarbazepine [51][52].
The way the results are presented can vary considerably from one report to another. The results can be delivered either as raw genetic data or as the ultimate therapeutic recommendation. In the reports, a proprietary iconography can be used for the description of results. This iconography can use specific symbols to indicate patients in which an increased risk of adverse effects or therapy ineffectiveness is expected. Other reports can use a traffic light view with three main drug categories: green for normal risk, yellow for use with caution, and red for exclusion. Results displayed in any format can cause the provider to oversimplify the PGx test results, ignoring additional clinical considerations.
Below (Fig. 1) is an example of an abbreviated PGx test performed by AyassBioScience
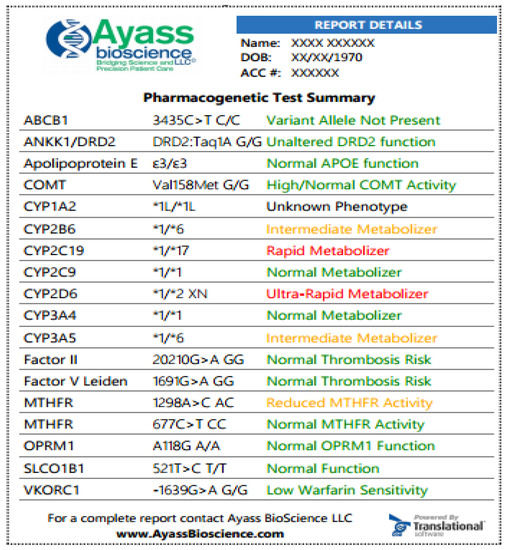
Fig. 1. Samples of PGx test at AyassBioScience: The report is featured with color-coded information for easier navigation and attention. The greed color indicates that the medication can be prescribed according to standard regimens, and the risk for the indicated condition is not increased; yellow color—indicated that dosage adjusting is required, there is an increased vigilance or the patient has a moderate risk for the indicated condition; red color designates that medication has potentially reduced efficacy and increased toxicity or the patient has an increased risk for the indicated condition. In the exemplified results, the patient has a normal response to Apixaban (the drug is a substrate for the efflux transport proteins P-gp (ABCB1) and BCRP (ABCG2) and, possibly, decreased response to Bupropion. Bupropion is metabolized to its active metabolite hydroxybupropion by CYP2B6. This metabolite contributes to the therapeutic effects of bupropion when used as a smoking cessation agent or as an antidepressant. The patient has also increased response to Codeine, which is converted into its active metabolite morphine by CYP2D6. Since this patient is the ultra-rapid metabolizer (UM), a greatly increased morphine level is expected, and the patient is at high risk of toxicity when taking codeine.
A variety of ways of presenting the information can be used in the report. Since each way is different, one should be careful to ensure a complete understanding of the meaning of each categorization [43].
2.4. Automation Tools for Integrating PGx Testing into the Clinic
Rapidly developing new technologies of DNA sequencing enabled quick and efficient identification of the genomic characteristics of organisms. The main result of the genomic and post-genomic technologies development was a significant expansion of the capabilities to study the genetic nature of a whole spectrum of human diseases. A genome-wide association study (GWAS) of clinical samples generated data on the genetic makeup featured for the specific groups (families or populations) to elaborate a personalized treatment approach. In this regard, to date, the research into the mechanisms of genetic predisposition to multifactorial diseases and the identification of specific genetic markers are of particular relevance. Such methods are widely used internationally and in Russia, where modern sequencing technologies are gradually introduced into medical research and medical practice to personify the treatment strategy.
Next-generation sequencing (NGS) is used for in-depth (multiple) reading of genetic material, which is necessary, for instance, for re-sequencing and assembly of new genomes (de novo), transcriptome, and epigenomic studies. This method allows one to reveal rare variations and to understand the genetic function better. However, the avalanche of new data will also make problems for researchers and clinicians, giving many “options of unknown importance” in the absence of clear indications [43]. Modifications of CDS are required to ensure the storage and use of new data architecture and new data availability programs. This, in turn, will identify significant opportunities in the coming years. Several healthcare systems use CDS tools to integrate PGx test data into clinical decision-making and provide information to end-users [53]. CDS systems can be used to administer high-risk drugs and provide automated recommendations indicating why certain modifications should be applied to a selected drug or dose.
The U-PGx PREPARE study developed solutions for sites with limited electronic health record infrastructure. The “Safety-Code” card is part of a mobile CDS, and with a quick response code, a medical professional is directed to a website with dosing recommendations customized for the patient [54]. This card also provides an overview of the most relevant PGx test results with a list of drugs with existing (known) recommendations [55].
Such CDS tools will be necessary, as PGx tests become more common due to the emergence of new results and test formats. One can also focus on developing patient-centered applications and portals, through which the patient can interact with his or her service providers and receive a consultation based on outcomes.
The increase in providers’ and patients’ awareness can stimulate the use of PGx testing. Consequently, laboratories will adjust the scope and type of available clinical PGx tests based on clinical requirements. The expected increase in the development of multigene panel PGx tests follows the advances in oncology, microbiology, and other fields. However, the proper clinical use of such tests appears to be more involved, requiring the support and participation of multiple interested parties.
3. Conclusions
We are on the verge of a new era in human genetic analysis. Deciphering the human genome, together with the development of high-throughput genetic analysis methods, provided a unique opportunity for the identification of complex genetic changes, resulting in the development of new branches of pharmacy: pharmacogenetics and pharmacogenomics. The genomic profiling of patients became a new diagnostic tool, enabling personalized drug therapy with higher effectiveness and fewer adverse effects. The pharmacogenetic tests can help select a specific drug with a specific dosage and administration regime, which will meet the requirements for the treatment of a particular patient in a specific setting, allowing one to avoid time-consuming dose adjustment inevitably associated with adverse effects.
Although PGx testing represents a significant therapeutic advance, it is just a healthcare professional’s arsenal tool of the future, increasing therapy effectiveness. The technological breakthrough will never override a physician’s experience and logic. This is a small paradigm shift towards the personalization of treatment.
PGx testing development inevitably will stimulate the progress in methods of data storage and data analysis, which are required for the integration of modern information technologies into routine clinical practice. However, the improvement in digital technologies, the increase in the volume and accessibility of databases is not the only problem in integrating genetic testing into the healthcare continuum. This, in turn, requires changes in the interaction between an individual patient and the healthcare system since the ultimate goal is the patient’s recovery or control of the disease, regardless of what laboratory techniques and data analysis technologies are employed. In other words, the treatment should take into account both the patient’s requirements and the test results. Today pharmacogenetics is in its infancy. A large pool of experimental, but mostly pilot, studies in this area have been accumulated; however, this information can barely be classified as systemic, which makes it difficult to explain the observed correlations between the presence of polymorphisms of a separate gene and epigenetic factors, or the severity of the course of the disease/resistance to therapy. The authors do not aspect information on congenital/acquired polymorphisms, even for the genetically determined diseases, such as cardiovascular diseases and diabetes mellitus. So far, pharmacogenetics provides mosaic information related to the association between the response to drug therapy depending on the genetic background. The next stage is expected to be the research on a larger group of participants, study the contribution of epigenetic factors, and providing clinical guidelines for adjusting or selecting the therapy based on the personal characteristics of the patient. Nevertheless, the field of pharmacogenetics is being actively developed and discussed, and the current demand for end-products in medicine is unusually high. We expect that soon researchers find answers to many questions that are still controversial.
References
- Alonso, S.G.; de la Torre Díez, I.; Zapiraín, B.G. Predictive, Personalized, Preventive and Participatory (4P) Medicine Applied to Telemedicine and EHealth in the Literature. J. Med. Syst. 2019, 43, 140.
- Vogel, F. Moderne Probleme der Humangenetik. In Ergebnisse der Inneren Medizin und Kinderheilkunde; Heilmeyer, L., Schoen, R., de Rudder, B., Eds.; Springer: Berlin/Heidelberg, Germany, 1959; pp. 52–125.
- Meyer, U.A. Pharmacogenetics—Five Decades of Therapeutic Lessons from Genetic Diversity. Nat. Rev. Genet. 2004, 5, 669–676.
- Caldwell, J. Drug Metabolism and Pharmacogenetics: The British Contribution to Fields of International Significance. Br. J. Pharmacol. 2006, 147 (Suppl. 1), S89–S99.
- Hassan, R.; Allali, I.; Agamah, F.E.; Elsheikh, S.S.M.; Thomford, N.E.; Dandara, C.; Chimusa, E.R. Drug Response in Association with Pharmacogenomics and Pharmacomicrobiomics: Towards a Better Personalized Medicine. Brief. Bioinform. 2020, bbaa292.
- Testa, R.; Bonfigli, A.R.; Sirolla, C.; Boemi, M.; Manfrini, S.; Mari, D.; Testa, I.; Sacchi, E.; Franceschi, C. Effect of 4G/5G PAI-1 Polymorphism on the Response of PAI-1 Activity to Vitamin E Supplementation in Type 2 Diabetic Patients. Diabetes Nutr. Metab. 2004, 17, 217–221.
- He, H.-Y.; Liu, M.-Z.; Zhang, Y.-L.; Zhang, W. Vitamin Pharmacogenomics: New Insight into Individual Differences in Diseases and Drug Responses. Genom. Proteom. Bioinform. 2017, 15, 94–100.
- Awh, C.C.; Lane, A.-M.; Hawken, S.; Zanke, B.; Kim, I.K. CFH and ARMS2 Genetic Polymorphisms Predict Response to Antioxidants and Zinc in Patients with Age-Related Macular Degeneration. Ophthalmology 2013, 120, 2317–2323.
- Mosolov, S.N. (Ed.) Biological Methods of Therapy for Mental Disorders; Sociopolitical thought (Socialno-politicheskaya mysl): Moscow, Russia, 2012. (In Russian)
- Fabbri, C.; Zohar, J.; Serretti, A. Pharmacogenetic Tests to Guide Drug Treatment in Depression: Comparison of the Available Testing Kits and Clinical Trials. Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 86, 36–44.
- Carter, C.A.; Frischmeyer-Guerrerio, P.A. The Genetics of Food Allergy. Curr. Allergy Asthma Rep. 2018, 18, 2.
- Cardon, L.R.; Harris, T. Precision Medicine, Genomics and Drug Discovery. Hum. Mol. Genet. 2016, 25, R166–R172.
- Marx, V. The DNA of a Nation. Nature 2015, 524, 503–505.
- Lu, M.; Lewis, C.M.; Traylor, M. Pharmacogenetic testing through the direct-to-consumer genetic testing company 23andMe. BMC Med. Genomics 2017, 10, 47.
- Crews, K.R.; Hicks, J.K.; Pui, C.-H.; Relling, M.V.; Evans, W.E. Pharmacogenomics and Individualized Medicine: Translating Science into Practice. Clin. Pharmacol. Ther. 2012, 92, 467–475.
- Veenstra, D.L. The Value of Routine Pharmacogenomic Screening—Are We There yet? A Perspective on the Costs and Benefits of Routine Screening—Shouldn’t Everyone Have This Done? Clin. Pharmacol. Ther. 2016, 99, 164–166.
- Haga, S.B.; Kantor, A. Horizon Scan of Clinical Laboratories Offering Pharmacogenetic Testing. Health Aff. 2018, 37, 717–723.
- de Leon, J.; Susce, M.T.; Murray-Carmichael, E. The AmpliChip CYP450 Genotyping Test: Integrating a New Clinical Tool. Mol. Diagn. Ther. 2006, 10, 135–151.
- Burmester, J.K.; Sedova, M.; Shapero, M.H.; Mansfield, E. DMET Microarray Technology for Pharmacogenomics-Based Personalized Medicine. Methods Mol. Biol. 2010, 632, 99–124.
- Johnson, J.A.; Burkley, B.M.; Langaee, T.Y.; Clare-Salzler, M.J.; Klein, T.E.; Altman, R.B. Implementing Personalized Medicine: Development of a Cost-Effective Customized Pharmacogenetics Genotyping Array. Clin. Pharmacol. Ther. 2012, 92, 437–439.
- Bielinski, S.J.; Olson, J.E.; Pathak, J.; Weinshilboum, R.M.; Wang, L.; Lyke, K.J.; Ryu, E.; Targonski, P.V.; Van Norstrand, M.D.; Hathcock, M.A.; et al. Preemptive Genotyping for Personalized Medicine: Design of the Right Drug, Right Dose, Right Time-Using Genomic Data to Individualize Treatment Protocol. Mayo Clin. Proc. 2014, 89, 25–33.
- Chambers, C.; Jansen, L.A.; Dhamija, R. Review of Commercially Available Epilepsy Genetic Panels. J. Genet. Couns. 2016, 25, 213–217.
- Platt, J.; Cox, R.; Enns, G.M. Points to Consider in the Clinical Use of NGS Panels for Mitochondrial Disease: An Analysis of Gene Inclusion and Consent Forms. J. Genet. Couns. 2014, 23, 594–603.
- Bush, W.S.; Crosslin, D.R.; Owusu-Obeng, A.; Wallace, J.; Almoguera, B.; Basford, M.A.; Bielinski, S.J.; Carrell, D.S.; Connolly, J.J.; Crawford, D.; et al. Genetic Variation among 82 Pharmacogenes: The PGRNseq Data from the EMERGE Network. Clin. Pharmacol. Ther. 2016, 100, 160–169.
- Janssens, A.C.J.W.; Deverka, P.A. Useless Until Proven Effective: The Clinical Utility of Preemptive Pharmacogenetic Testing. Clin. Pharmacol. Ther. 2014, 96, 652–654.
- Lazaridis, K.N. Improving Therapeutic Odyssey: Preemptive Pharmacogenomics Utility in Patient Care. Clin. Pharmacol. Ther. 2017, 101, 39–41.
- Dunnenberger, H.M.; Crews, K.R.; Hoffman, J.M.; Caudle, K.E.; Broeckel, U.; Howard, S.C.; Hunkler, R.J.; Klein, T.E.; Evans, W.E.; Relling, M.V. Preemptive Clinical Pharmacogenetics Implementation: Current Programs in Five US Medical Centers. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 89–106.
- Hicks, J.K.; Sangkuhl, K.; Swen, J.J.; Ellingrod, V.L.; Müller, D.J.; Shimoda, K.; Bishop, J.R.; Kharasch, E.D.; Skaar, T.C.; Gaedigk, A.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2D6 and CYP2C19 Genotypes and Dosing of Tricyclic Antidepressants: 2016 Update. Clin. Pharmacol. Ther. 2017, 102, 37–44.
- Hicks, J.K.; Bishop, J.R.; Sangkuhl, K.; Müller, D.J.; Ji, Y.; Leckband, S.G.; Leeder, J.S.; Graham, R.L.; Chiulli, D.L.; LLerena, A.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin. Pharmacol. Ther. 2015, 98, 127–134.
- Vilches, S.; Tuson, M.; Vieta, E.; Álvarez, E.; Espadaler, J. Effectiveness of a Pharmacogenetic Tool at Improving Treatment Efficacy in Major Depressive Disorder: A Meta-Analysis of Three Clinical Studies. Pharmaceutics 2019, 11, 453.
- Frazer, A.; Benmansour, S. Delayed Pharmacological Effects of Antidepressants. Mol. Psychiatry 2002, 7 (Suppl. 1), S23–S28.
- GlaxoSmithKline Inc. ZIAGEN (GlaxoSmithKline Inc): FDA Package Insert. Available online: https://druginserts.com/lib/rx/meds/ziagen-6/ (accessed on 24 August 2020).
- Cho, S.-M.; Lee, K.-Y.; Choi, J.R.; Lee, K.-A. Development and Comparison of Warfarin Dosing Algorithms in Stroke Patients. Yonsei Med. J. 2016, 57, 635–640.
- Kelly, L.E.; Rieder, M.; van den Anker, J.; Malkin, B.; Ross, C.; Neely, M.N.; Carleton, B.; Hayden, M.R.; Madadi, P.; Koren, G. More Codeine Fatalities after Tonsillectomy in North American Children. Pediatrics 2012, 129, e1343–e1347.
- Thorn, C.F.; Klein, T.E.; Altman, R.B. Codeine and Morphine Pathway. Pharm. Genom. 2009, 19, 556–558.
- Crews, K.R.; Gaedigk, A.; Dunnenberger, H.M.; Klein, T.E.; Shen, D.D.; Callaghan, J.T.; Kharasch, E.D.; Skaar, T.C. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for Codeine Therapy in the Context of Cytochrome P450 2D6 (CYP2D6) Genotype. Clin. Pharmacol. Ther. 2012, 91, 321–326.
- Lemke, A.A.; Hulick, P.J.; Wake, D.T.; Wang, C.; Sereika, A.W.; Yu, K.D.; Glaser, N.S.; Dunnenberger, H.M. Patient Perspectives Following Pharmacogenomics Results Disclosure in an Integrated Health System. Pharmacogenomics 2018, 19, 321–331.
- Patel, H.N.; Ursan, I.D.; Zueger, P.M.; Cavallari, L.H.; Pickard, A.S. Stakeholder Views on Pharmacogenomic Testing. Pharmacotherapy 2014, 34, 151–165.
- Haga, S.B.; Mills, R.; Moaddeb, J.; Allen Lapointe, N.; Cho, A.; Ginsburg, G.S. Patient Experiences with Pharmacogenetic Testing in a Primary Care Setting. Pharmacogenomics 2016, 17, 1629–1636.
- Bielinski, S.J.; St Sauver, J.L.; Olson, J.E.; Wieland, M.L.; Vitek, C.R.; Bell, E.J.; Mc Gree, M.E.; Jacobson, D.J.; McCormick, J.B.; Takahashi, P.Y.; et al. Are Patients Willing to Incur Out-of-Pocket Costs for Pharmacogenomic Testing? Pharm. J. 2017, 17, 1–3.
- Haga, S.B.; O’Daniel, J.M.; Tindall, G.M.; Lipkus, I.R.; Agans, R. Survey of US Public Attitudes toward Pharmacogenetic Testing. Pharm. J. 2012, 12, 197–204.
- Dunnenberger, H.M.; Biszewski, M.; Bell, G.C.; Sereika, A.; May, H.; Johnson, S.G.; Hulick, P.J.; Khandekar, J. Implementation of a Multidisciplinary Pharmacogenomics Clinic in a Community Health System. Am. J. Health Syst. Pharm. 2016, 73, 1956–1966.
- Wake, D.T.; Ilbawi, N.; Dunnenberger, H.M.; Hulick, P.J. Pharmacogenomics. Med. Clin. N. Am. 2019, 103, 977–990.
- Scott, S.A.; Sangkuhl, K.; Stein, C.M.; Hulot, J.-S.; Mega, J.L.; Roden, D.M.; Klein, T.E.; Sabatine, M.S.; Johnson, J.A.; Shuldiner, A.R. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C19 Genotype and Clopidogrel Therapy: 2013 Update. Clin. Pharmacol. Ther. 2013, 94, 317–323.
- Vo, T.T.; Bell, G.C.; Obeng, A.O.; Hicks, J.K.; Dunnenberger, H.M. Pharmacogenomics Implementation: Considerations for Selecting a Reference Laboratory. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2017, 37, 1014–1022.
- Bousman, C.; Maruf, A.A.; Müller, D.J. Towards the Integration of Pharmacogenetics in Psychiatry: A Minimum, Evidence-Based Genetic Testing Panel. Curr. Opin. Psychiatry 2019, 32, 7–15.
- Phillips, E.J.; Sukasem, C.; Whirl-Carrillo, M.; Müller, D.J.; Dunnenberger, H.M.; Chantratita, W.; Goldspiel, B.; Chen, Y.-T.; Carleton, B.C.; George, A.L.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for HLA Genotype and Use of Carbamazepine and Oxcarbazepine: 2017 Update. Clin. Pharmacol. Ther. 2018, 103, 574–581.
- Hosono, N.; Kato, M.; Kiyotani, K.; Mushiroda, T.; Takata, S.; Sato, H.; Amitani, H.; Tsuchiya, Y.; Yamazaki, K.; Tsunoda, T.; et al. CYP2D6 Genotyping for Functional-Gene Dosage Analysis by Allele Copy Number Detection. Clin. Chem. 2009, 55, 1546–1554.
- Caudle, K.E.; Dunnenberger, H.M.; Freimuth, R.R.; Peterson, J.F.; Burlison, J.D.; Whirl-Carrillo, M.; Scott, S.A.; Rehm, H.L.; Williams, M.S.; Klein, T.E.; et al. Standardizing Terms for Clinical Pharmacogenetic Test Results: Consensus Terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet. Med. 2017, 19, 215–223.
- Luzum, J.A.; Pakyz, R.E.; Elsey, A.R.; Haidar, C.E.; Peterson, J.F.; Whirl-Carrillo, M.; Handelman, S.K.; Palmer, K.; Pulley, J.M.; Beller, M.; et al. Pharmacogenomics Research Network Translational Pharmacogenetics Program. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Outcomes and Metrics of Pharmacogenetic Implementations Across Diverse Healthcare Systems. Clin. Pharmacol. Ther. 2017, 102, 502–510.
- Hershfield, M.S.; Callaghan, J.T.; Tassaneeyakul, W.; Mushiroda, T.; Thorn, C.F.; Klein, T.E.; Lee, M.T.M. Clinical Pharmacogenetics Implementation Consortium Guidelines for Human Leukocyte Antigen-B Genotype and Allopurinol Dosing. Clin. Pharmacol. Ther. 2013, 93, 153–158.
- Leckband, S.G.; Kelsoe, J.R.; Dunnenberger, H.M.; George, A.L.; Tran, E.; Berger, R.; Müller, D.J.; Whirl-Carrillo, M.; Caudle, K.E.; Pirmohamed, M. Clinical Pharmacogenetics Implementation Consortium Guidelines for HLA-B Genotype and Carbamazepine Dosing. Clin. Pharmacol. Ther. 2013, 94, 324–328.
- Hinderer, M.; Boeker, M.; Wagner, S.A.; Lablans, M.; Newe, S.; Hülsemann, J.L.; Neumaier, M.; Binder, H.; Renz, H.; Acker, T.; et al. Integrating Clinical Decision Support Systems for Pharmacogenomic Testing into Clinical Routine—A Scoping Review of Designs of User-System Interactions in Recent System Development. BMC Med. Inform. Decis. Mak. 2017, 17, 81.
- Blagec, K.; Koopmann, R.; Crommentuijn-van Rhenen, M.; Holsappel, I.; van der Wouden, C.H.; Konta, L.; Xu, H.; Steinberger, D.; Just, E.; Swen, J.J.; et al. Implementing Pharmacogenomics Decision Support across Seven European Countries: The Ubiquitous Pharmacogenomics (U-PGx) Project. J. Am. Med. Inform. Assoc 2018, 25, 893–898.
- Krebs, K.; Milani, L. Translating Pharmacogenomics into Clinical Decisions: Do Not Let the Perfect Be the Enemy of the Good. Hum. Genom. 2019, 13, 39.




