
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Bing He | -- | 3365 | 2023-12-19 09:12:11 | | | |
| 2 | Lindsay Dong | Meta information modification | 3365 | 2023-12-22 01:34:54 | | |
Video Upload Options
Oilseed crops are rich in plant lipids that not only provide essential fatty acids for the human diet but also play important roles as major sources of biofuels and indispensable raw materials for the chemical industry. The regulation of lipid metabolism genes is a major factor affecting oil production.
1. Introduction
Function of Lipids
Classification of Plant Lipids
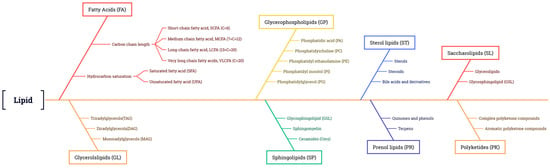
Lipid Accumulation and Storage in Plants
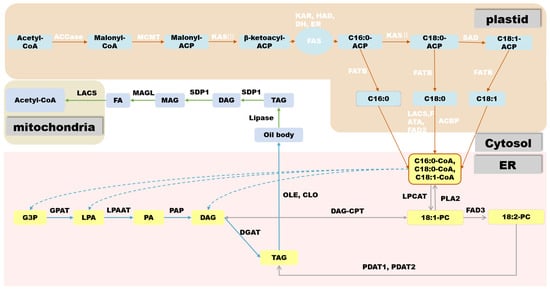
In oilseed crops, the first step in FA biosynthesis is the conversion of acetyl-CoA produced via sucrose glycolysis to malonyl-CoA by acetyl-CoA carboxylase (ACCase). ACCase is a type I biotin-containing enzyme and there are two main forms of ACCase in plants: Heteromeric ACCaseI and Homomeric ACCaseII. ACCase is composed of four subunits: biotin carboxylase (BC), biotin carboxyl carrier protein (BCCP), α-carboxyltransferase (α-CT), and β-carboxyltransferase (β-CT). It has three functional domains, namely the BC functional domain, the BCCP functional domain, and the CT functional domain, respectively [10][11]. After its biosynthesis, malonyl-CoA is transferred to the ACP component of the fatty acid synthase (FAS) complex by malonyl-CoA:ACP malonyltransacylase (MCMT). In plants, FAS is a multi-component type-II enzyme located in the plastids, consisting of 3-β-ketoacyl-ACP synthase III (KASIII), β-ketoacyl-ACP synthase (KASI), ketoacyl-ACP reductase (KAR), hydroxyacyl-ACP reductase (HAD), and enoyl-ACP reductase (ENR). FAS uses acetyl-CoA as the starting unit for a condensation reaction. Each elongation cycle is supplied with a two-carbon unit by malonyl-ACP to produce 16:0-ACP and 18:0-ACP after seven or eight cycles, respectively, at which point 18:0-ACP passes through Δ9 stearoyl-ACP desaturase to form 18:1-ACP, making phosphatidic acid (PA) (16:0) and oleic acid (18:1) the major products of FA biosynthesis in most plant plastids [12].
In most oilseed crops, such as soybean (Glycine max) and rapeseed (Brassica napus), TAGs are stored in seeds as an energy source for germination and aid in seed dispersal [13]. TAG consists of a glycerol backbone and three FA molecules chemically linked by ester bonds, providing a carbon skeleton and energy source. TAG biosynthesis and accumulation occur through a complex network of reactions taking place in the plastid, cytoplasm, and ER. Depending on the plant species, TAG can accumulate in different organs, mainly in embryonic tissues (rapeseed) or endosperm tissues (castor bean). In oilseeds that store oil in the embryo, the main storage tissue is the cotyledons, but substantial seed oil can also accumulate in the hypocotyl, radicle, and surrounding endosperm/aleurone layers [14]. After acyl-CoA is transported from the plastid to the ER, the most common pathway for TAG biosynthesis is the acyl-CoA-dependent Kennedy pathway [15]. In this pathway, acyl-CoA is incorporated into glycerol-3-phosphate (G3P) by acyl-CoA:glycerol-3-phosphate acyltransferase (GPAT) and lysophosphatidic acid acyltransferase (LPAT) at the sn-1 and sn-2 positions of G3P, respectively, to form PA.
Cytoplasmic lipid droplets (LDs) are organelles that store non-polar lipids such as TAGs and sterol esters [16]. In mature seeds, LDs are distributed in the central region of storage cells and are mostly oval or irregular in shape [17]. The primary LD structure consists of a phospholipid monolayer coated with various proteins. The current general model of LD biogenesis is that non-poplar lipids such as TAGs are first produced by membrane-associated enzymes in the ER and then accumulate in the form of a lens between lobes of the ER membrane, culminating with LD formation on the cytoplasmic side of the ER membrane [18]. Two important proteins have recently been shown to be involved in LD formation: SEIPIN [19][20] and lipid-droplet-associated protein in Arabidopsis (Arabidopsis thaliana) [21].
2. Progress in the Identification of Key Genes behind Lipid Metabolism in Plants
2.1. Identification and Functional Characterization of Key Genes
2.2. Transcription Factors Involved in Regulation
2.3. Advances in Multi-Omics Studies of Oilseed Crops
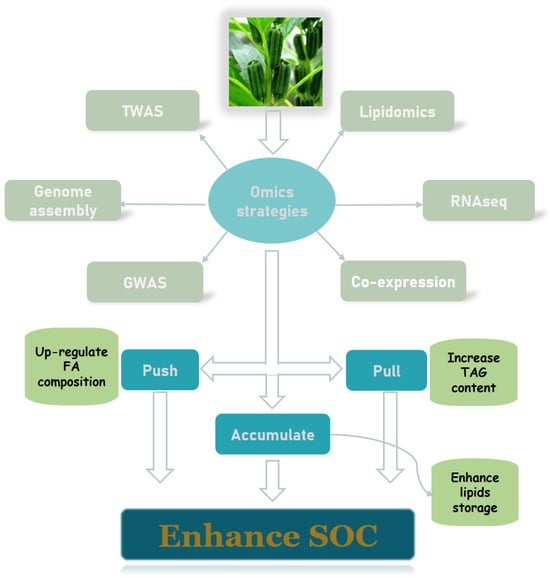
2.3.1. De Novo Genome Sequencing and Annotation
2.3.2. Identification of Differentially Expressed Genes
2.3.3. Construction of Oil Co-Expression Networks
2.3.4. Genome-Wide Association Studies to Map Oil Content–Related Loci
2.3.5. Lipidomics for Oil Structure and Quality Identification
References
- Jorge, N.; Goncalves, L.A.G.; Dobarganes, M.C. Influence of fatty acid composition on the formation of polar glycerides and polar fatty acids in sunflower oils heated at frying temperatures. Grasas Aceites 1997, 48, 17–24.
- De Carvalho, C.; Caramujo, M. The Various Roles of Fatty Acids. Molecules 2018, 23, 2583.
- Slocombe, S.P.; Cornah, J.; Pinfield-Wells, H.; Soady, K.; Zhang, Q.Y.; Gilday, A.; Dyer, J.M.; Graham, I.A. Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol. J. 2009, 7, 694–703.
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytol. 2020, 227, 698–713.
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.; Glass, C.; Merrill, A.H.; Murphy, R.; Raetz, C.; Russell, D.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, D527–D532.
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–861.
- Ohlrogge, J.; Thrower, N.; Mhaske, V.; Stymne, S.; Baxter, M.; Yang, W.; Liu, J.; Shaw, K.; Shorrosh, B.; Zhang, M.; et al. PlantFAdb: A resource for exploring hundreds of plant fatty acid structures synthesized by thousands of plants and their phylogenetic relationships. Plant J. 2018, 96, 1299–1308.
- Harwood, J.L. Fatty Acid Metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988, 39, 101–138.
- Lunn, D.; Wallis, J.G.; Browse, J. A multigene approach secures hydroxy fatty acid production in Arabidopsis. J. Exp. Bot. 2021, 73, 2875–2888.
- Alban, C.; Baldet, P.; Douce, R. Localization and characterization of 2 structurally different forms of acetyl-coa carboxylase in young pea leaves, of which one is sensitive to aryloxyphenoxypropionate herbicides. Biochem. J. 1994, 300, 557–565.
- Kozaki, A.K.; Mayumi, K.; Sasaki, Y. Thiol-disulfide exchange between nuclear-encoded and chloroplast-encoded subunits of pea acetyl-CoA carboxylase. J. Biol. Chem. 2001, 276, 39919–39925.
- Bates, P.D. Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2016, 1861, 1214–1225.
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14.
- Lu, S.; Sturtevant, D.; Aziz, M.; Jin, C.; Li, Q.; Chapman, K.D.; Guo, L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J. 2018, 94, 915–932.
- Kennedy, E.P. Biosynthesis of complex lipids. Fed. Proc. 1961, 20, 934–940.
- Ischebeck, T.; Krawczyk, H.E.; Mullen, R.T.; Dyer, J.M.; Chapman, K.D. Lipid droplets in plants and algae: Distribution, formation, turnover and function. Semin. Cell Dev. Biol. 2020, 108, 82–93.
- Yin, D.; Wang, Y.; Zhang, X.; Li, H.; Lu, X.; Zhang, J.; Zhang, W.; Chen, S. De Novo Assembly of the Peanut (Arachis hypogaea L.) Seed Transcriptome Revealed Candidate Unigenes for Oil Accumulation Pathways. PLoS ONE 2013, 8, e73767.
- Thiam, A.R.; Ikonen, E. Lipid Droplet Nucleation. Trends Cell Biol. 2021, 31, 108–118.
- Taurino, M.; Costantini, S.; De Domenico, S.; Stefanelli, F.; Ruano, G.; Delgadillo, M.O.; Sanchez-Serrano, J.J.; Sanmartin, M.; Santino, A.; Rojo, E. SEIPIN Proteins Mediate Lipid Droplet Biogenesis to Promote Pollen Transmission and Reduce Seed Dormancy. Plant Physiol. 2018, 176, 1531–1546.
- Kelly, A.; Shaw, E.; Powers, S.; Kurup, S.; Eastmond, P. Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.). Plant Biotechnol J. 2013, 11, 355–361.
- Pyc, M.; Gidda, S.K.; Seay, D.; Esnay, N.; Kretzschmar, F.K.; Cai, Y.Q.; Doner, N.M.; Greer, M.S.; Hull, J.J.; Coulon, D.; et al. LDIP cooperates with SEIPIN and LDAP to facilitate lipid droplet biogenesis in Arabidopsis. Plant Cell 2021, 33, 3076–3103.
- Chia, T.Y.P.; Pike, M.J.; Rawsthorne, S. Storage oil breakdown during embryo development of Brassica napus (L.). J. Exp. Bot. 2005, 56, 1285–1296.
- Chiofalo, B.; Lo Presti, V. 404-Sampling Techniques for the Determination of Volatile Components in Food of Animal Origin. In Comprehensive Sampling Sample Preparation; Pawliszyn, J., Ed.; Academic Press: Oxford, UK, 2012; pp. 61–85.
- Huang, A.H.C. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996, 110, 1055–1061.
- Goepfert, S.; Poirier, Y. Beta-oxidation in fatty acid degradation and beyond. Curr. Opin. Plant Biol. 2007, 10, 245–251.
- Gupta, R.; Gupta, N. Lipid Biosynthesis and Degradation; Springer: Singapore, 2021; pp. 491–523.
- Ma, J.; Sun, S.; Whelan, J.; Shou, H. CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds. Int. J. Mol. Sci. 2021, 22, 3877.
- Raboanatahiry, N.; Chao, H.; Guo, L.; Gan, J.; Xiang, J.; Yan, M.; Zhang, L.; Yu, L.; Li, M. Synteny analysis of genes and distribution of loci controlling oil content and fatty acid profile based on QTL alignment map in Brassica napus. BMC Genom. 2017, 18, 776.
- Zou, J.; Katavic, V.; Giblin, E.M.; Barton, D.L.; Mackenzie, S.L.; Keller, W.A.; Hu, X.; Taylor, D.C. Modification of seed oil content and acyl composition in the brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell 1997, 9, 909–923.
- Chen, S.L.; Lei, Y.; Xu, X.; Huang, J.Q.; Jiang, H.F.; Wang, J.; Cheng, Z.S.; Zhang, J.A.; Song, Y.H.; Liao, B.S.; et al. The Peanut (Arachis hypogaea L.) Gene AhLPAT2 Increases the Lipid Content of Transgenic Arabidopsis Seeds. PLoS ONE 2015, 10, e0136170.
- Jako, C.; Kumar, A.; Wei, Y.; Zou, J.; Barton, D.L.; Giblin, E.M.; Covello, P.S.; Taylor, D.C. Seed-Specific Over-Expression of an Arabidopsis cDNA Encoding a Diacylglycerol Acyltransferase Enhances Seed Oil Content and Seed Weight. Plant Physiol. 2001, 126, 861–874.
- Wang, Z.K.; Huang, W.J.; Chang, J.M.; Sebastian, A.; Li, Y.G.; Li, H.Y.; Wu, X.X.; Zhang, B.B.; Meng, F.L.; Li, W.B. Overexpression of SiDGAT1, a gene encoding acyl-CoA:diacylglycerol acyltransferase from Sesamum indicum L. increases oil content in transgenic Arabidopsis and soybean. Plant Cell Tissue Organ Cult. 2014, 119, 399–410.
- Akoh, C.C.; Lee, G.C.; Liaw, Y.C.; Huang, T.H.; Shaw, J.F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004, 43, 534–552.
- Chen, M.X.; Du, X.; Zhu, Y.; Wang, Z.; Hua, S.J.; Li, Z.L.; Guo, W.L.; Zhang, G.P.; Peng, J.R.; Jiang, L.X. Seed Fatty Acid Reducer acts downstream of gibberellin signalling pathway to lower seed fatty acid storage in Arabidopsis. Plant Cell Environ. 2012, 35, 2155–2169.
- Brocard-Gifford, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 2003, 131, 78–92.
- Zhai, Z.Y.; Keereetaweep, J.; Liu, H.; Feil, R.; Lunn, J.E.; Shanklin, J. Trehalose 6-Phosphate Positively Regulates Fatty Acid Synthesis by Stabilizing WRINKLED1. Plant Cell 2018, 30, 2616–2627.
- Guo, Y.; Huang, Y.; Gao, J.; Pu, Y.; Wang, N.; Shen, W.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; et al. CIPK9 is involved in seed oil regulation in Brassica napus L. and Arabidopsis thaliana (L.) Heynh. Biotechnol. Biofuels 2018, 11, 124.
- Zafar, S.; Li, Y.-L.; Li, N.-N.; Zhu, K.-M.; Tan, X.-L. Recent advances in enhancement of oil content in oilseed crops. J. Biotechnol. 2019, 301, 35–44.
- Fei, W.J.; Yang, S.Q.; Hu, J.; Yang, F.; Qu, G.Y.; Peng, D.; Zhou, B. Research advances of WRINKLED1 (WRI1) in plants. Funct. Plant Biol. 2020, 47, 185–194.
- Benning, N.F.a.C. wrinkled1: A Novel, Low-Seed-Oil Mutant of Arabidopsis with a Deficiency in the Seed-Specific Regulation of Carbohydrate Metabolism. Plant Physiol. 1998, 118, 91–101.
- Ji, X.-J.; Mao, X.; Hao, Q.-T.; Liu, B.-L.; Xue, J.-A.; Li, R.-Z. Splice Variants of the Castor WRI1 Gene Upregulate Fatty Acid and Oil Biosynthesis When Expressed in Tobacco Leaves. Int. J. Mol. Sci. 2018, 19, 146.
- Mu, J.Y.; Tan, H.L.; Zheng, Q.; Fu, F.Y.; Liang, Y.; Zhang, J.; Yang, X.H.; Wang, T.; Chong, K.; Wang, X.J.; et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 1042–1054.
- Tan, H.; Yang, X.; Zhang, F.; Zheng, X.; Qu, C.; Mu, J.; Fu, F.; Li, J.; Guan, R.; Zhang, H.; et al. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 2011, 156, 1577–1588.
- Elahi, N.; Duncan, R.W.; Stasolla, C. Decreased seed oil production in FUSCA3 Brassica napus mutant plants. Plant Physiol. Biochem. 2015, 96, 222–230.
- Manan, S.; Zhao, J. Role of Glycine max ABSCISIC ACID INSENSITIVE 3 (GmABI3) in lipid biosynthesis and stress tolerance in soybean. Funct. Plant Biol. 2021, 48, 171.
- Pouvreau, B.; Blundell, C.; Vohra, H.; Zwart, A.B.; Arndell, T.; Singh, S.; Vanhercke, T. A Versatile High Throughput Screening Platform for Plant Metabolic Engineering Highlights the Major Role of ABI3 in Lipid Metabolism Regulation. Front. Plant Sci. 2020, 11, 288.
- Chen, Y.H.; Yang, X.Y.; He, K.; Liu, M.H.; Li, J.G.; Gao, Z.F.; Lin, Z.Q.; Zhang, Y.F.; Wang, X.X.; Qiu, X.M.; et al. The MYB transcription factor superfamily of arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124.
- To, A.; Joubes, J.; Thueux, J.; Kazaz, S.; Lepiniec, L.; Baud, S. AtMYB92 enhances fatty acid synthesis and suberin deposition in leaves of Nicotiana benthamiana. Plant J. 2020, 103, 660–676.
- Liu, J.; Hua, W.; Yang, H.L.; Zhan, G.M.; Li, R.J.; Deng, L.B.; Wang, X.F.; Liu, G.H.; Wang, H.Z. The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J. Exp. Bot. 2012, 63, 3727–3740.
- Libault, M.; Farmer, A.; Joshi, T.; Takahashi, K.; Langley, R.J.; Franklin, L.D.; He, J.; Xu, D.; May, G.; Stacey, G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010, 63, 86–99.
- Severin, A.J.; Woody, J.L.; Bolon, Y.-T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160.
- Huang, X.; Han, B. Natural Variations and Genome-Wide Association Studies in Crop Plants. Annu. Rev. Plant Biol. 2014, 65, 531–551.
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168.
- Lieberman-Aiden, E.; Van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293.
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953.
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.-C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876.
- Li, Y.-H.; Zhou, G.; Ma, J.; Jiang, W.; Jin, L.-G.; Zhang, Z.; Guo, Y.; Zhang, J.; Sui, Y.; Zheng, L.; et al. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 2014, 32, 1045–1052.
- Liu, Y.C.; Du, H.L.; Li, P.C.; Shen, Y.T.; Peng, H.; Liu, S.L.; Zhou, G.A.; Zhang, H.K.; Liu, Z.; Shi, M.; et al. Pan-Genome of Wild and Cultivated Soybeans. Cell 2020, 182, 162–176.
- Zhang, L.; Wang, S.-B.; Li, Q.-G.; Song, J.; Hao, Y.-Q.; Zhou, L.; Zheng, H.-Q.; Dunwell, J.M.; Zhang, Y.-M. An Integrated Bioinformatics Analysis Reveals Divergent Evolutionary Pattern of Oil Biosynthesis in High- and Low-Oil Plants. PLoS ONE 2016, 11, e0154882.
- Wang, J.; Singh, S.K.; Du, C.; Li, C.; Fan, J.; Pattanaik, S.; Yuan, L. Comparative Transcriptomic Analysis of Two Brassica napus Near-Isogenic Lines Reveals a Network of Genes That Influences Seed Oil Accumulation. Front. Plant Sci. 2016, 7, 1498.
- O’Rourke, J.A.; Bolon, Y.T.; Bucciarelli, B.; Vance, C.P. Legume genomics: Understanding biology through DNA and RNA sequencing. Ann. Bot. 2014, 113, 1107–1120.
- Yang, S.; Miao, L.; He, J.; Zhang, K.; Li, Y.; Gai, J. Dynamic Transcriptome Changes Related to Oil Accumulation in Developing Soybean Seeds. Int. J. Mol. Sci. 2019, 20, 2202.
- Huang, R.; Zhou, Y.; Zhang, J.; Ji, F.; Jin, F.; Fan, W.; Pei, D. Transcriptome Analysis of Walnut (Juglans regia L.) Embryos Reveals Key Developmental Stages and Genes Involved in Lipid Biosynthesis and Polyunsaturated Fatty Acid Metabolism. J. Agric. Food Chem. 2021, 69, 377–396.
- Liu, J.; Dong, L.; Duan, R.; Hu, L.; Zhao, Y.; Zhang, L.; Wang, X. Transcriptomic Analysis Reveals the Regulatory Networks and Hub Genes Controlling the Unsaturated Fatty Acid Contents of Developing Seed in Soybean. Front. Plant Sci. 2022, 13, 876371.
- Niu, Y.; Wu, L.; Li, Y.; Huang, H.; Qian, M.; Sun, W.; Zhu, H.; Xu, Y.; Fan, Y.; Mahmood, U.; et al. Deciphering the transcriptional regulatory networks that control size, color, and oil content in Brassica rapa seeds. Biotechnol. Biofuels 2020, 13, 90.
- Zhang, Z.; Dunwell, J.M.; Zhang, Y.M. An integrated omics analysis reveals molecular mechanisms that are associated with differences in seed oil content between Glycine max and Brassica napus. BMC Plant Biol. 2018, 18, 328.
- Yao, M.; Guan, M.; Zhang, Z.; Zhang, Q.; Cui, Y.; Chen, H.; Liu, W.; Jan, H.U.; Voss-Fels, K.P.; Werner, C.R.; et al. GWAS and co-expression network combination uncovers multigenes with close linkage effects on the oleic acid content accumulation in Brassica napus. BMC Genom. 2020, 21, 320.
- Guerin, C.; Joët, T.; Serret, J.; Lashermes, P.; Vaissayre, V.; Agbessi, M.D.T.; Beulé, T.; Severac, D.; Amblard, P.; Tregear, J.; et al. Gene coexpression network analysis of oil biosynthesis in an interspecific backcross of oil palm. Plant J. 2016, 87, 423–441.
- Flint-Garcia, S.A.; Thuillet, A.-C.; Yu, J.; Pressoir, G.; Romero, S.M.; Mitchell, S.E.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 2005, 44, 1054–1064.
- Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 725.
- Borisjuk, L.; Neuberger, T.; Schwender, J.; Heinzel, N.; Sunderhaus, S.; Fuchs, J.; Hay, J.O.; Tschiersch, H.; Braun, H.P.; Denolf, P.; et al. Seed architecture shapes embryo metabolism in oilseed rape. Plant Cell 2013, 25, 1625–1640.
- Zhang, G.; Ahmad, M.Z.; Chen, B.; Manan, S.; Zhang, Y.; Jin, H.; Wang, X.; Zhao, J. Lipidomic and transcriptomic profiling of developing nodules reveals the essential roles of active glycolysis and fatty acid and membrane lipid biosynthesis in soybean nodulation. Plant J. 2020, 103, 1351–1371.
- Zhang, D.; Guo, X.; Wang, Q.; Zhao, L.Y.; Sun, Q.C.; Duan, X.L.; Cao, Y.P.; Sun, H. Investigation on lipid profile of peanut oil and changes during roasting by lipidomic approach. Lwt-Food Sci. Technol. 2022, 154, 112594.
- Shi, T.; Wu, G.; Jin, Q.; Wang, X. Detection of camellia oil adulteration using chemometrics based on fatty acids GC fingerprints and phytosterols GC-MS fingerprints. Food Chem. 2021, 352, 129422.
- Huang, Y.; Ma, R.; Xu, Y.; Zhong, K.; Bu, Q.; Gao, H. A Comparison of Lipid Contents in Different Types of Peanut Cultivars Using UPLC-Q-TOF-MS-Based Lipidomic Study. Foods 2021, 11, 4.
- Demiral, I.; Dogan, M.; Bastu, E.; Buyru, F. Genomic, proteomic and lipidomic evaluation of endometrial receptivity. Turk. J. Obstet. Gynecol. 2015, 12, 237–243.




