Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Dacheng Wei | -- | 1929 | 2023-11-06 08:52:55 | | | |
| 2 | Rita Xu | Meta information modification | 1929 | 2023-11-06 09:22:16 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Liang, Q.; Cao, B.; Xiao, Q.; Wei, D. Graphene Field-Effect Transistor Biosensors in COVID-19 Detection Technology. Encyclopedia. Available online: https://encyclopedia.pub/entry/51174 (accessed on 07 February 2026).
Liang Q, Cao B, Xiao Q, Wei D. Graphene Field-Effect Transistor Biosensors in COVID-19 Detection Technology. Encyclopedia. Available at: https://encyclopedia.pub/entry/51174. Accessed February 07, 2026.
Liang, Qin-Hong, Ban-Peng Cao, Qiang Xiao, Dacheng Wei. "Graphene Field-Effect Transistor Biosensors in COVID-19 Detection Technology" Encyclopedia, https://encyclopedia.pub/entry/51174 (accessed February 07, 2026).
Liang, Q., Cao, B., Xiao, Q., & Wei, D. (2023, November 06). Graphene Field-Effect Transistor Biosensors in COVID-19 Detection Technology. In Encyclopedia. https://encyclopedia.pub/entry/51174
Liang, Qin-Hong, et al. "Graphene Field-Effect Transistor Biosensors in COVID-19 Detection Technology." Encyclopedia. Web. 06 November, 2023.
Copy Citation
Coronavirus disease 2019 (COVID-19) is a disease caused by the infectious agent of severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2). Graphene field-effect transistor (GFET) biosensors have become the most promising diagnostic technology for detecting SARS-CoV-2 due to their advantages of high sensitivity, fast-detection speed, label-free operation, and low detection limit.
GFET biosensor
COVID-19
biological sensors
1. Introduction
In 2019, a novel coronavirus as a kind of severe acute respiratory syndrome coronavirus (SARS-CoV-2) swept the world, causing a respiratory disease which is named coronavirus disease 2019 (COVID-19) [1][2]. COVID-19 is reportedly transmitted by airborne and contact-transmission respiratory droplets [3][4]. The methods for diagnosing COVID-19 can be roughly divided into three categories [5]: chest CT scanning [6], serological testing [7][8], and nucleic acid testing [9]. Chest CT scanning is limited to recognizing the virus type and is only available in hospitals. Serological testing is not suitable for early diagnosis of infection because the number of antibodies in our bodies gradually increases for at least a week after being infected with COVID-19 [10][11][12]. The nucleic acid detection method requires skilled professionals, and it takes 4–6 h to obtain the results [13]. Real-time reverse transcription-polymerase chain reaction (RT-PCR) remains the gold standard for COVID-19 diagnosis. However, RT-PCR requires a gene amplification process, tedious preparation steps, expensive equipment, specialized laboratories, and technicians, which reduces the efficiency of the test [14]. Therefore, developing a real-time, fast, accurate, easy-to-operate, and low-cost diagnostic technology is one of the key challenges in the fight against COVID-19.
Biosensors with simple operation and rapid detection have been widely considered a superior alternative detection technology [15]. In recent years, an endless stream of biosensors has been studied [16]. There are many types of biosensors, such as field-effect transistor (FET) biosensors [17], optical biosensors [18], plasmon resonance biosensors [19], and electrochemical biosensors [20]. These biosensors have become a powerful new means of detecting various biomolecules for diagnostics. In particular, GFET biosensors have the advantages of high sensitivity, fast detection speeds, no labels, and low detection limits and have become the most promising technology for COVID-19 detection [21][22]. GFET biosensors detect SARS-CoV-2 through specific targets on the surface of the graphene channel. SARS-CoV-2 is a segmented, enveloped coronavirus family with a single-stranded RNA structure [23]. The genome inside the particle encodes four structural proteins. Viral RNA is coated with nucleocapsid (N) protein. In addition to nucleocapsid (N) proteins, there are spike proteins (S), membrane proteins (M), and envelope proteins (E), which are embedded in the lipid bilayer [24]. Therefore, SARS-CoV-2 can be detected by using nucleic acid molecules or structural proteins as targets for the specific reaction (Figure 1). Currently, GFET biosensors for detecting COVID-19 mainly use nucleic acid hybridization or antigen–antibody-specific reaction for detection.
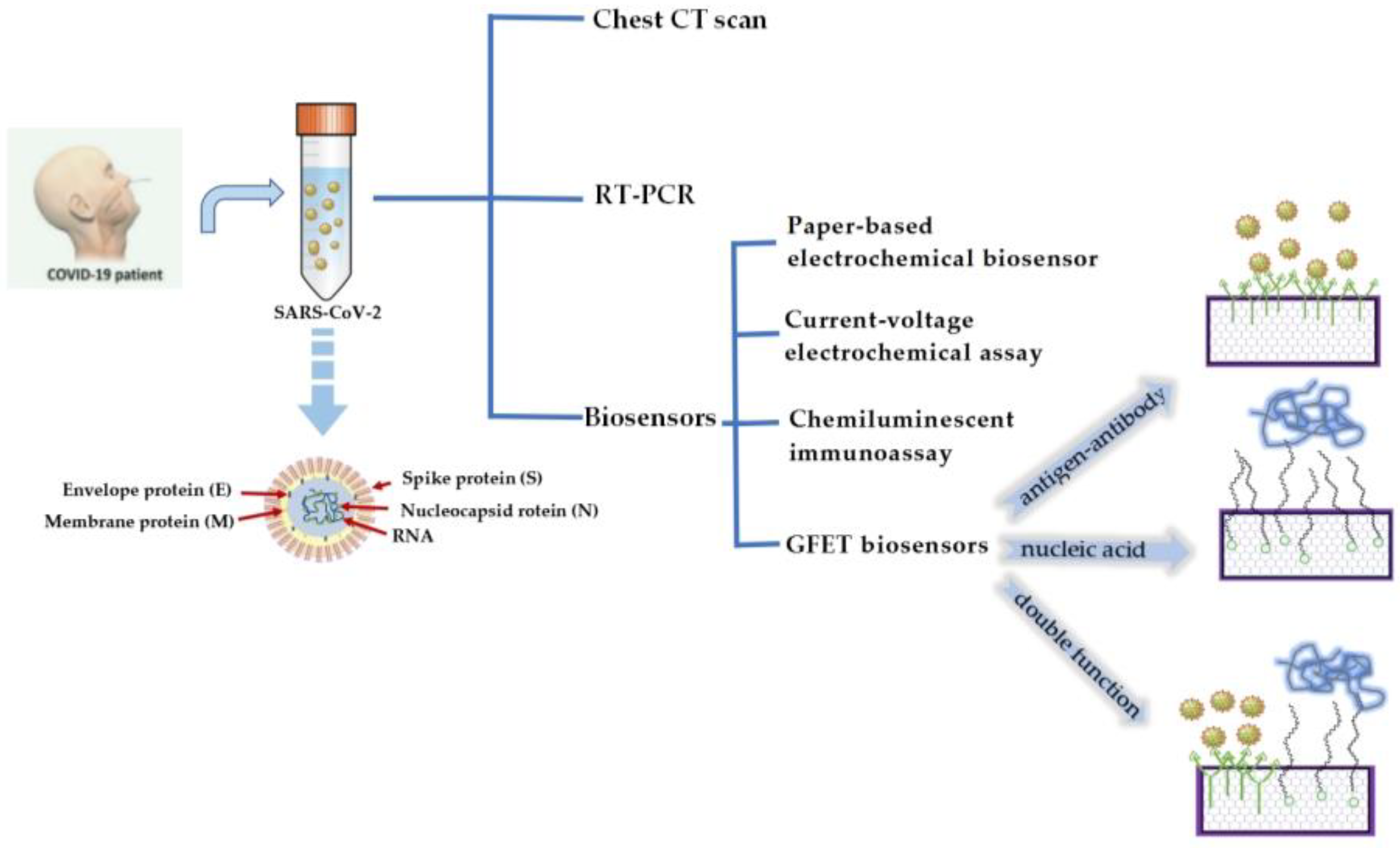
Figure 1. Several detection methods for COVID-19 and the schematic diagram of a GFET biosensor for detecting SARS-CoV-2.
2. Application of GFET Biosensors in the Diagnosis of COVID-19
GFET biosensors have great potential for diagnosing and controlling disease transmission in COVID-19 and other biomolecular detections. Compared with traditional detection technology, GFET biosensors have certain advantages in the bedside maintenance of biomolecules because of their excellent performance.
2.1. GFET Biosensors Detect SARS-CoV-2 Based on Specific Antigen–Antibody Binding
The coronavirus S protein is a large, multifunctional transmembrane fusion glycoprotein of the class I virus. The S protein is attached to the surface of the viral particle and determines the shape of the virus’ crown-like appearance. The coronavirus N protein promotes the assembly of viral particle and plays a role in the formation of the viral genome. After being infected with SARS-CoV-2, B lymphocytes or B cells produce five types of antibodies, IgA, IgG, IgM, IgD, and IgE, known as immunoglobulins [10]. Therefore, many researchers have detected SARS-CoV-2 by targeting antibodies. An ultra-sensitive GFET biosensor was prepared by Wei’s group [25]. The SARS-CoV-2 spike S1 protein was modified on the surface of the sensor to detect the spike S1 antibody (Figure 2). The SARS-CoV-2 spike S1 protein was immobilized on the surface of the graphene channel to realize biological functionalization. The strong specific binding between antibodies and proteins affected the concentration of the graphene channel medium and obtained a measurable electrical response.
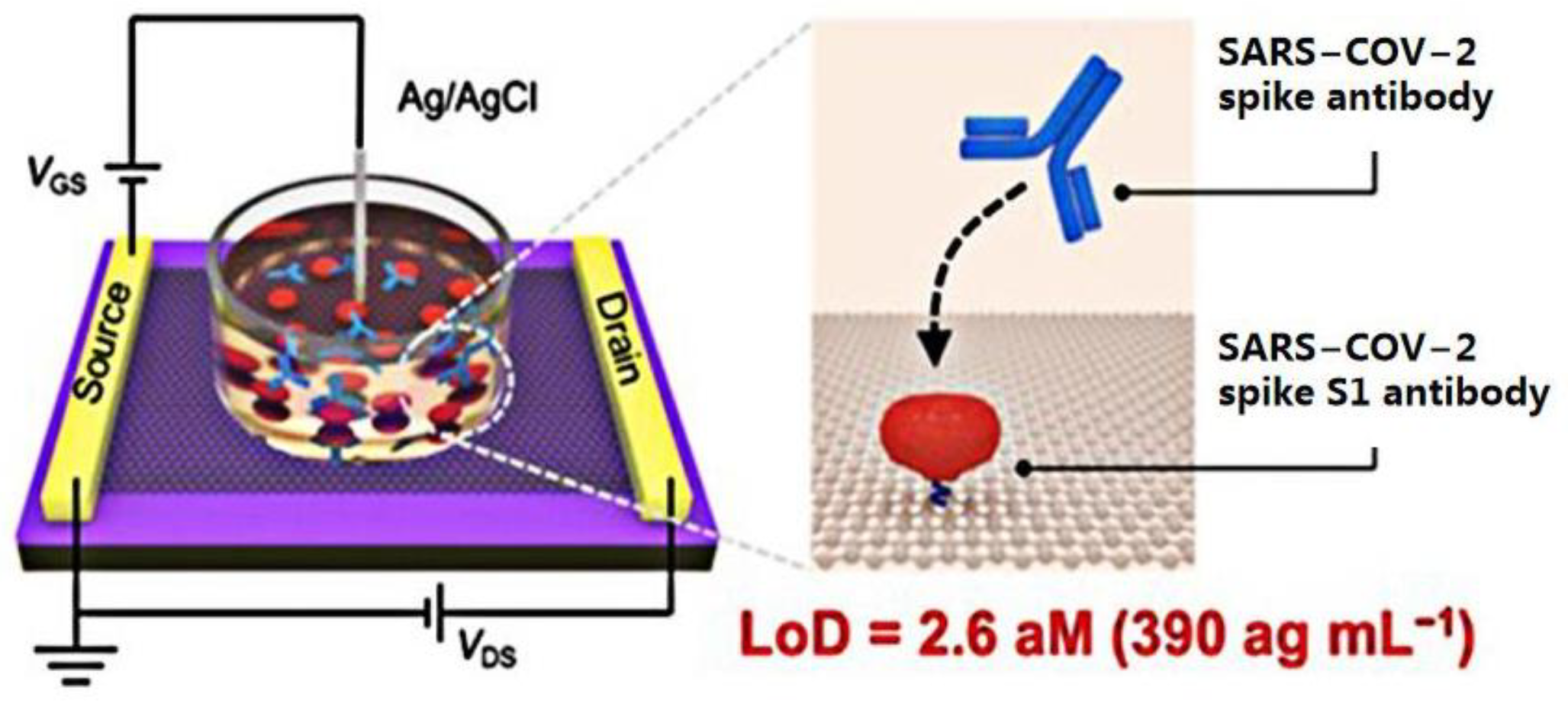
Figure 2. Schematic diagram of the GFET biosensor for detecting SARS-CoV-2 spike antibodies.
In recent years, laser-induced graphene technology has become a new way of preparing GFET biosensors. Cui’s group [26] first used lasers to manufacture a graphene channel, using a 450 nm UV laser of 840 MW, and the electrode region was manufactured using a UV laser of 900 MW. The graphene channel region obtained after laser radiation showed a spongy porous shape, which expanded its binding area with biomolecules. This research group has developed a one-step, simple, sensitive, and suitable method for the large-scale preparation of a laser-induced GFET biosensor. The SARS-CoV-2 spike antibody immobilized in graphene channels achieved rapid detection of the SARS-CoV-2 spike protein in 15 min at a detection limit of 1 pg/mL in phosphate-buffered saline (PBS) and 1 ng/mL in human serum, with high specificity for the target virus (Figure 3).
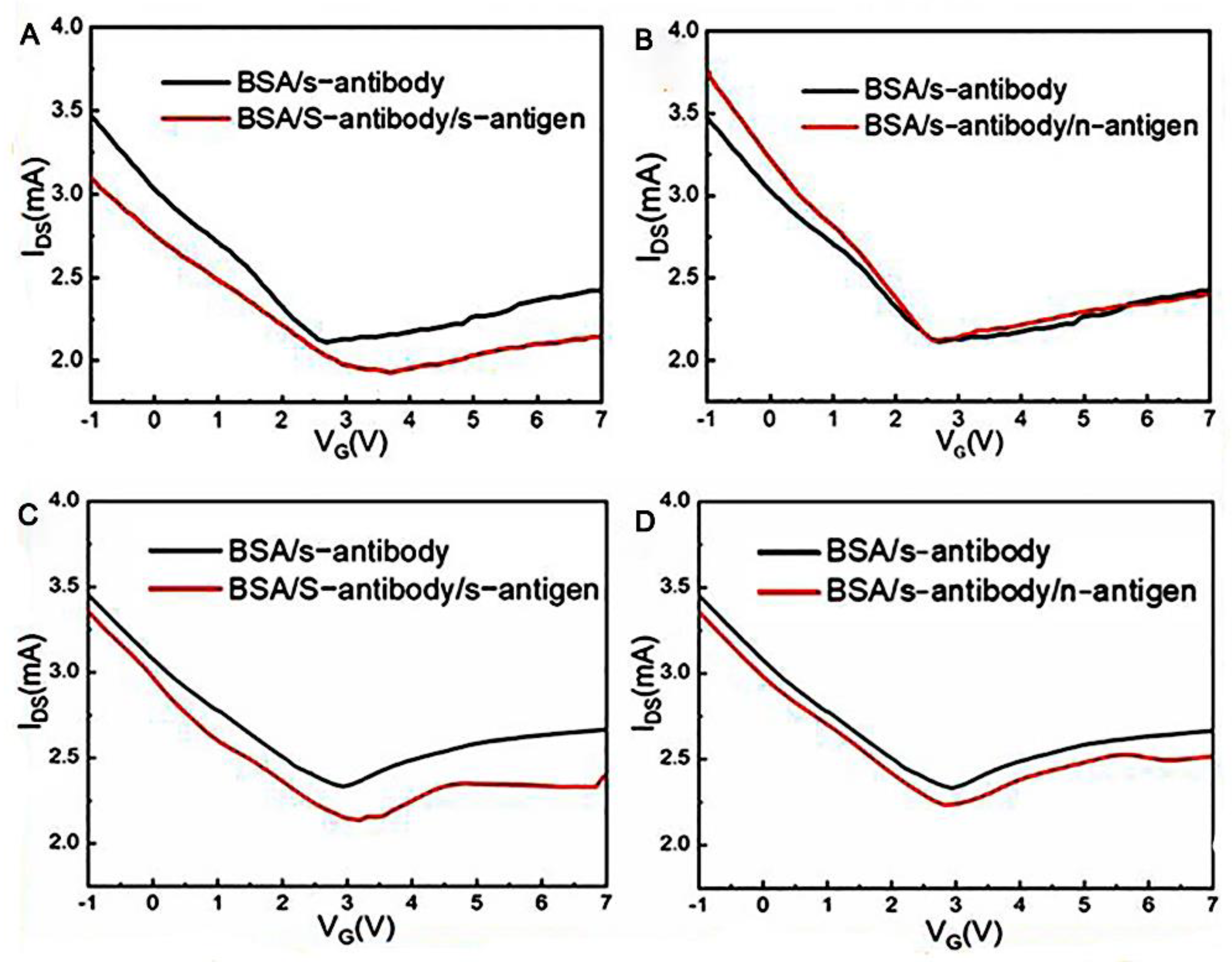
Figure 3. The virus detection performance of the laser-induced GFET. (A) Transfer characteristics of the laser-induced GFET biosensor responding to the complementary 1 pg/mL spike protein in PBS solution and (B) responding to the noncomplementary 1 pg/mL nucleocapsid protein in PBS solution. (C) Transfer characteristics of the laser-induced GFET biosensor responding to 1 pg/mL of complementary spike protein in human serum and (D) responding to 1 pg/mL noncomplementary nucleocapsid protein in human serum.
Shahdeo’s group [21] prepared GFET sensors on SiO2/Si substrates using transparent tape. The internally generated SARS-CoV-2 spike S1 antibody was immobilized on the carboxylic acid-activated graphene surface. The change in resistance generated by antibody–antigen interaction was monitored in response to the assay results (Figure 4). The results indicated that the device has high sensitivity and specificity and detects SARS-CoV-2 spike S1 proteins under conditions with a detection limit as low as 10 fM.
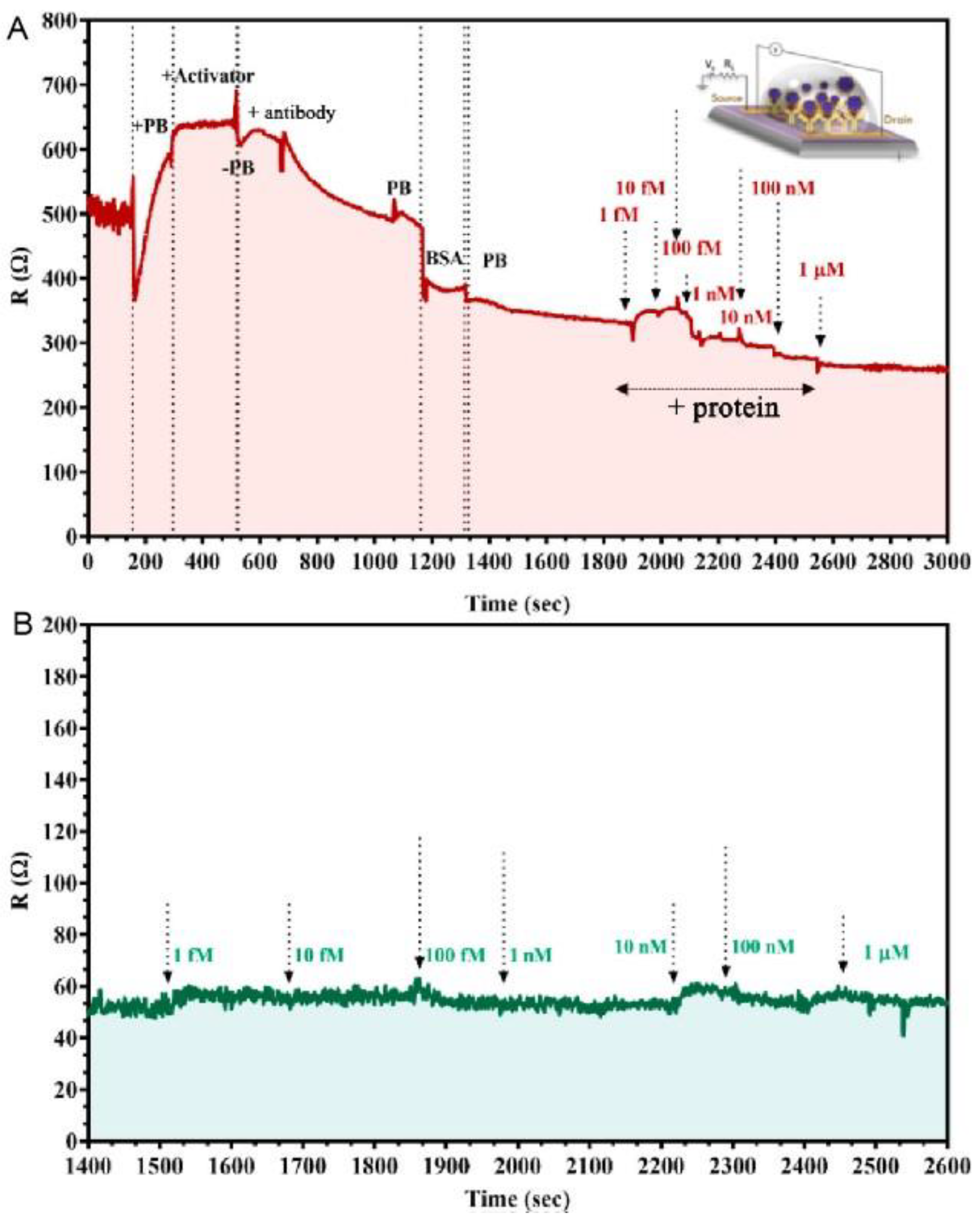
Figure 4. The kinetic response of the GFET device functionalized with the SARS-CoV-2 spike antibody at various concentrations of (A) SARS-CoV-2 spike protein added, ranging from 1 fM to 1 μM in 50 mM phosphate buffer (PB) (pH 7.2) and (B) MERS-CoV protein of various concentrations added (1 fM to 1 μM) in PB.
2.2. GFET Biosensors Based on Nucleic Acid Hybridization Detection of SARS-CoV-2
Nucleic acid detection is the most sensitive detection method for early viral infection and plays a key role in diagnosing and treating disease [27]. The RT-PCR method is the gold standard for detecting SARS-CoV-2, but the diagnostic process is complicated and time consuming. Therefore, many scholars tend to develop methods based on nucleic acid as a probe for detecting SARS-CoV-2. How to improve the sensitivity of the probe is an important problem. A lot of work has been carried out, mainly focused on the design of the probe and the development of sensing materials and new sensing mechanisms. In bioassays, different configurations are designed to improve the binding affinity with the target.
In recent years, the development of structural DNA nanotechnology has provided an accurate and controllable method for synthesizing various DNA nanostructures with specific functions. Different DNA nanostructures have different properties for biosensors. Compared with the single-probe nucleic acid hybridization detection method, the two recognition sites of dual probes can improve the sensitivity of virus detection. Wei’s group [28] developed a direct acid nucleic assay using a GFET with Y-shaped DNA dual probes (Figure 5). These Y-shaped DNA dual probe GFET biosensor could simultaneously identify ORF1ab and N gene regions, improving the sensitivity for identifying SARS-CoV-2. The synergistic effect of the two recognition sites of Y-shaped DNA dual probes improved the combination of DNA dual probes and targets. Therefore, the Y-type dual-probe GFET biosensor with an average of 40 s for response speed had excellent performance in terms of diagnosis time and detection limit.
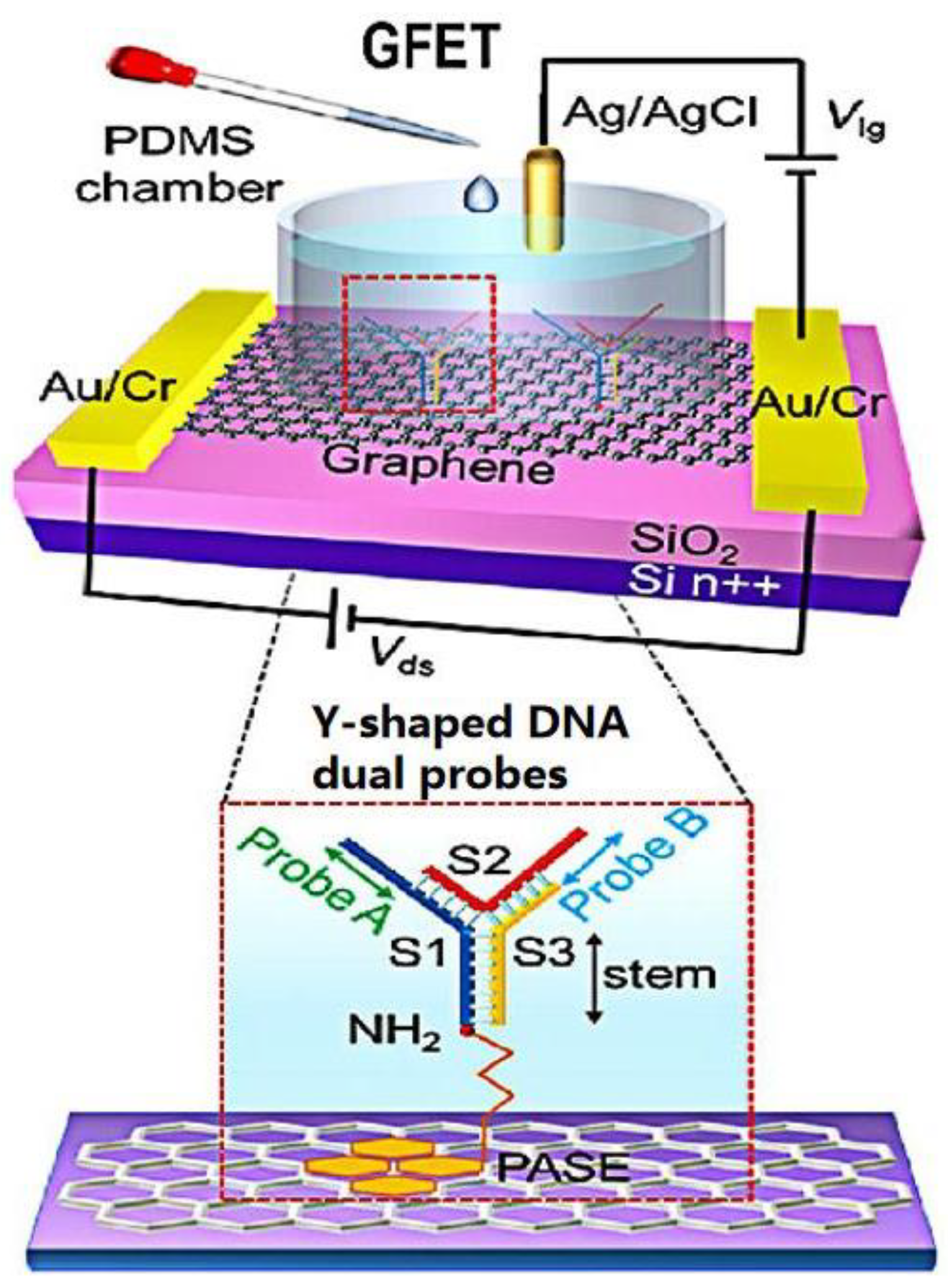
Figure 5. Schematic diagram of a Y-dual probe GFET biosensor. The dotted box is the structural schematic diagram of Y-shaped DNA dual probes.
Graphene surface-modified probe recognition sites greatly impact the performance of GFET biosensors. Therefore, improving the structural design of the probe is an important factor. Creatures with a multi-tentacle structure have strong olfactory sensitivity and capture and hunting ability. Inspired by these organisms, the sensitivity of multi-probe sensors can often be improved. Wei’s team designed the DNA nanostructure as a probe-tunable TDF dimer. The synergistic action of three probes improves the binding affinity and the sensitivity of the GFET biosensor. Wei’s group modified the GFET biosensor with a triple-probe tetrahedral DNA framework (TDF) to study its detection performance for SARS-CoV-2 RNA [29]. The triple-probe TDF dimer was modified on the surface of the graphene channel to form reaction targets (Figure 6). The sensor had highly specific recognition of RNA in the SARS-CoV-2 ORF1ab gene, RdRp gene, and E gene regions. As shown in Figure 7, under the same conditions, the response of the triple-probe TDF dimer was faster than that of dual-probe and single-probe TDF dimer sensors. The sensor identified all 14 positive cases in 30 nasopharyngeal swabs within an average diagnosis time of 74 s, showing promising prospects for real-time and centralized detection screening.
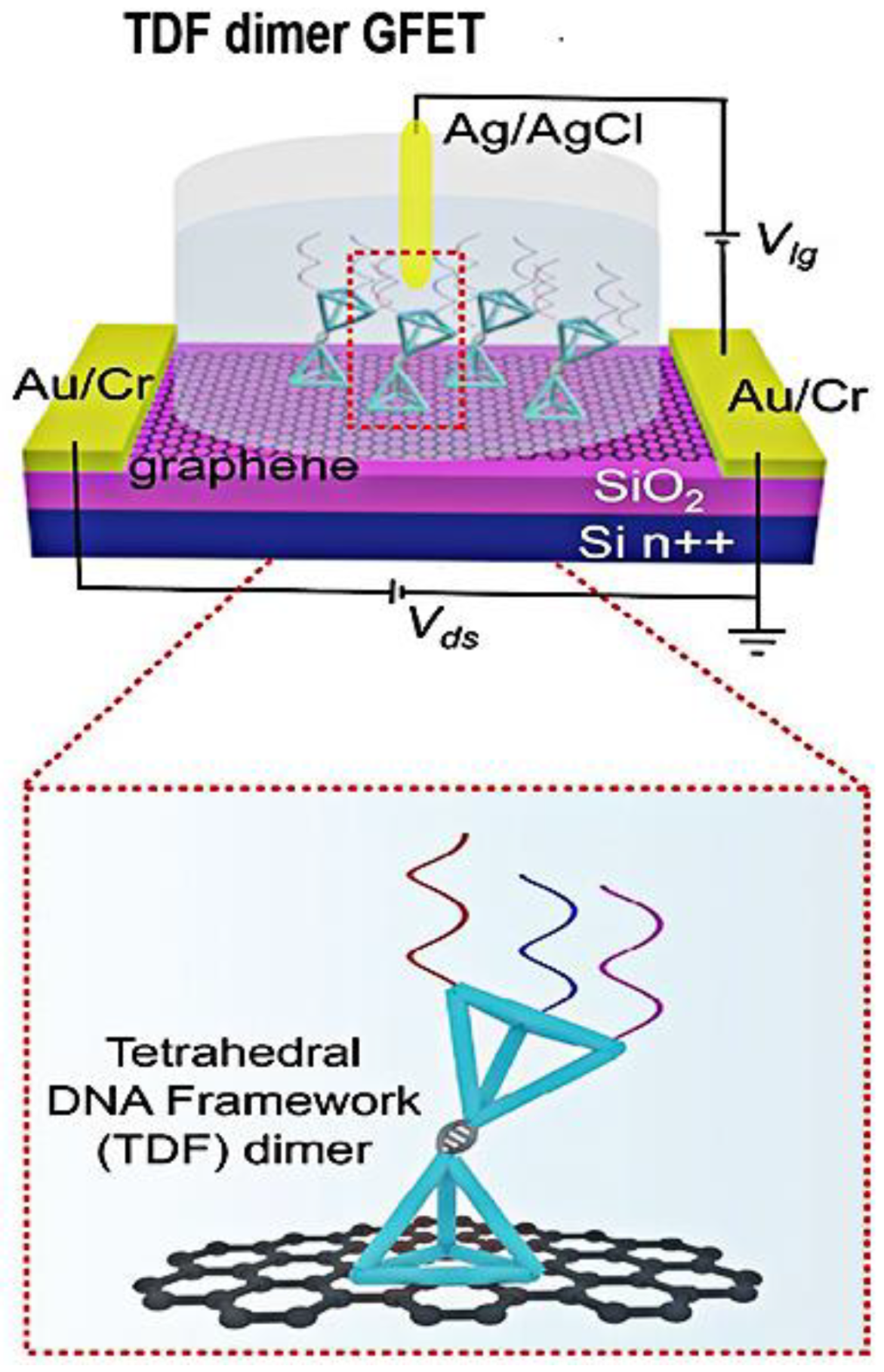
Figure 6. Schematic diagram of the triple-probe TDF dimer GFET sensor for SARS-CoV-2 RNA testing. The dotted box is the structural schematic diagram of TDF dimer.
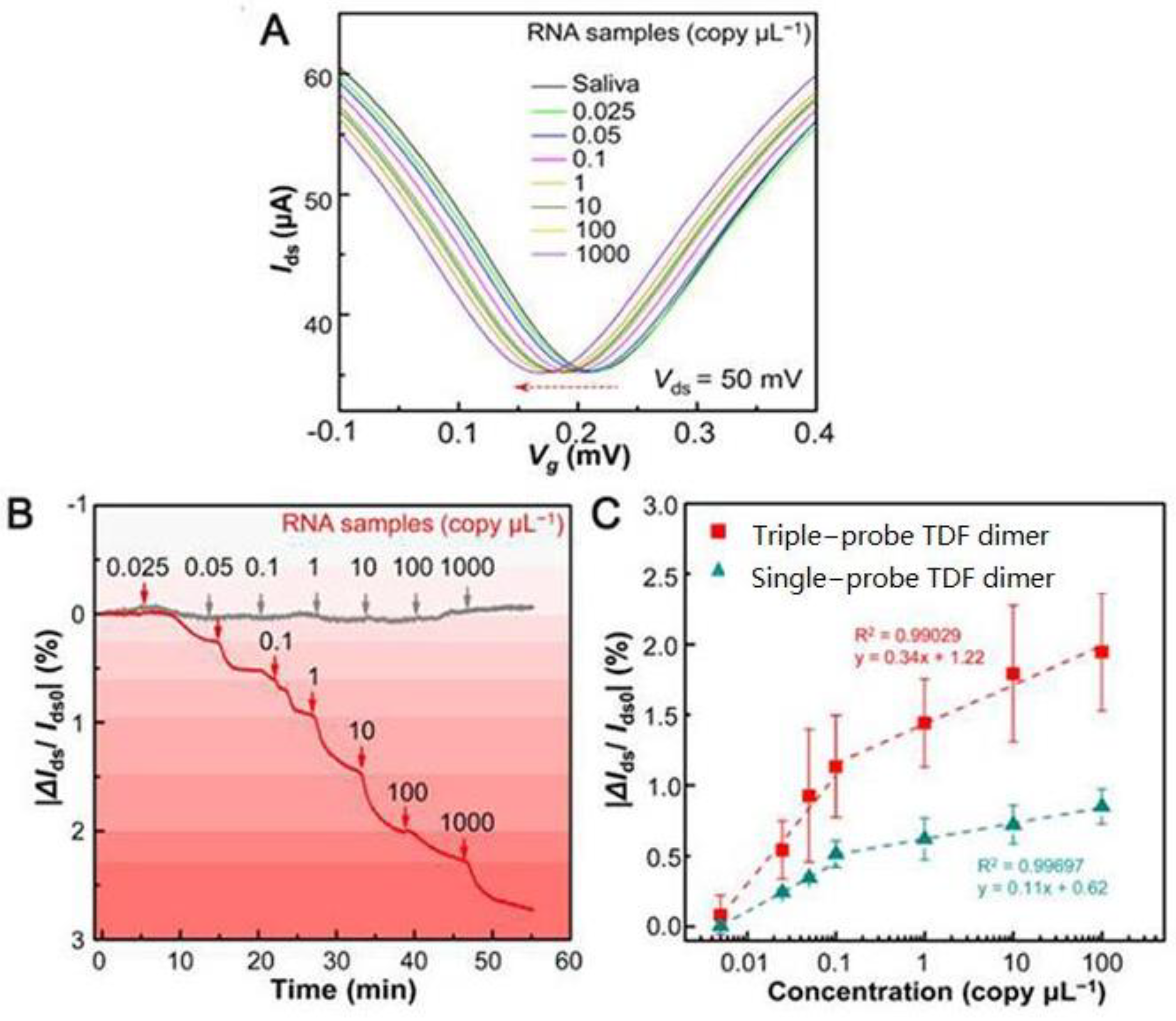
Figure 7. SARS-CoV-2 RNA testing. (A) Transfer curve measurement of adding different concentrations of target RNA (Ids−Vg response curve). (B) Real-time |ΔIds/Ids0| response upon different concentrations of target RNA (red line, modified with triple-probe TDF dimer; gray line, without immobilized probes). (C) |ΔIds/Ids0| responses of single- and triple-probe TDF dimer GFET sensors to different concentrations of target RNA.
2.3. Double Function of GFET Biosensors in Response to the Detection of SARS-CoV-2
Outstanding achievements have been made in detecting SARS-CoV-2 by single-response nucleic acid hybridization and antigen–antibody-specific reactions. The dual response of GFET biosensors to detecting SARS-CoV-2 has also received attention. Ke ’s group [30] reported a highly sensitive, specific, and convenient bi-functional GFET biosensor for detecting SARS-CoV-2 with detection limits as low as ~0.1 and ~1 fg·mL−1. The research group immobilized the SS-DNA probe or SARS-CoV-2 antigen protein on the surface of the graphene channel through π-π interaction. Detection results could be obtained in 5–10 min using SS-DNA probe-specific hybridization with a viral RNA polymerase target or SARS-CoV-2 antigen–antibody-specific recognition to convert biochemical effects into electrical signals. In order to verify the sensitivity and accuracy of the sensor for COVID-19 diagnosis, 18 volunteers were recruited for nucleic acid detection and 9 were recruited volunteers for immune detection.
Hwang’s group [31] was able to achieve high sensitivity by optimizing the crumpling ratio of the graphene sensing film. The results show that the crumpled GFET biosensor designed by Hwang’s group obtained good sensitivity and high reproducibility at a crumpling rate of about 55% [32]. The SARS-CoV-2 spike protein antibody and nucleocapsid protein antibody were immobilized on the surface of the graphene channel by π-π stacking, which could diagnose these two SARS-CoV-2 proteins at a lower detection limit. A field-effect transistor based on graphene oxide/graphene van der Waals heterostructures (GO/Gr heterostructure FET) was developed by Gao’s group [33]. Graphene oxide had abundant functional groups on its surface, and graphene oxide was superimposed onto graphene by π-π stacking, which enhances SARS-CoV-2 spike and nucleoprotein adsorption, improving the detection sensitivity of the sensor. The GO/Gr heterostructure FET sensor detects the SARS-CoV-2 protein in the range of 10 to 100 pg/mL with a limit detection as low as ~8 fg/mL. Meanwhile, as shown in Figure 8, the experimental data show ~3 × sensitivity enhancement compared with the GFET biosensor, which indicates its great potential for practical references in diagnosing SARS-CoV-2.
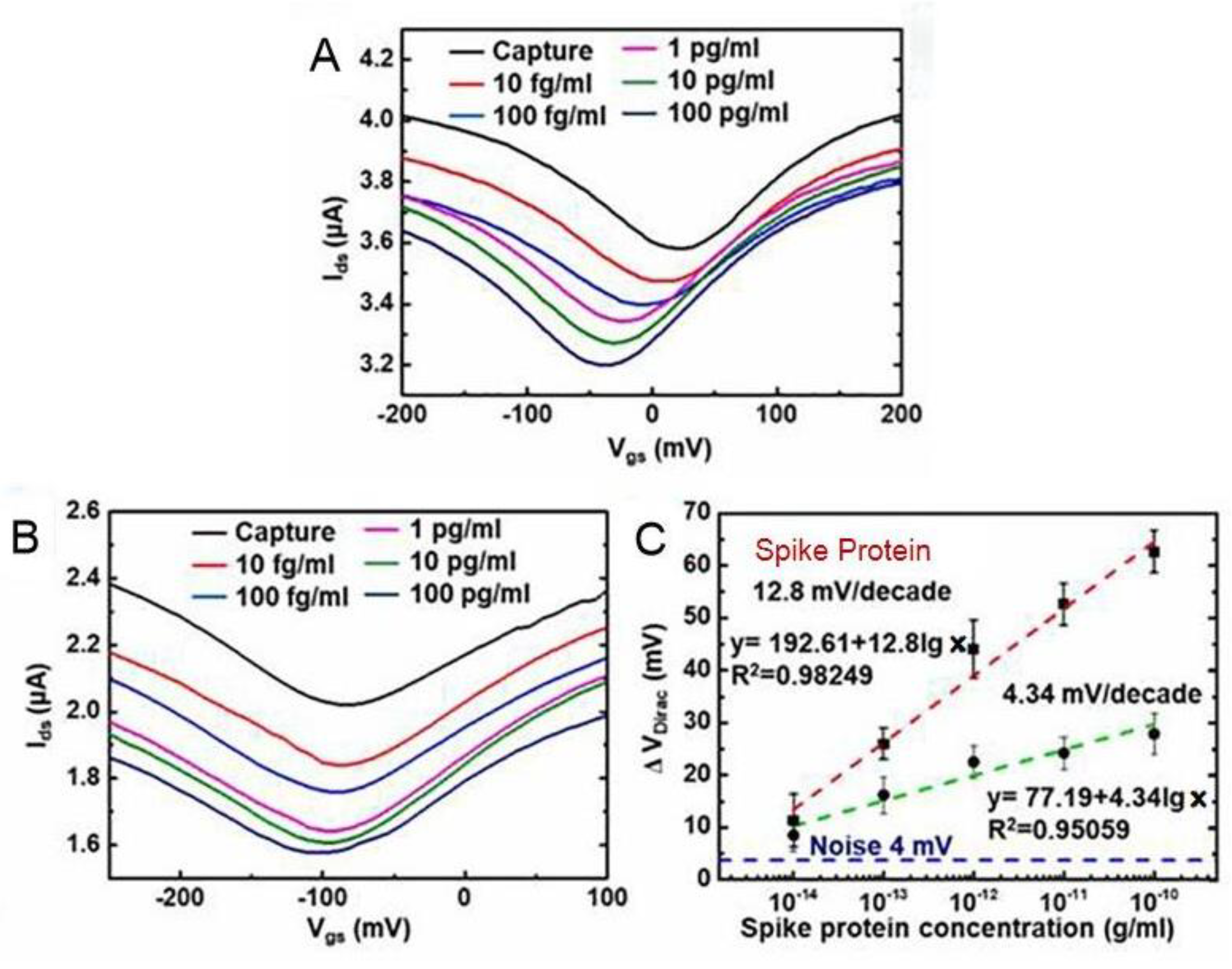
Figure 8. The SARS-CoV-2 spike protein concentrations dependent transfer curves of (A) GO/Gr FET biosensor and (B) Gr FET biosensor. (C) The SARS-CoV-2 spike protein concentrations dependent ΔVDirac shifts for both GO/Gr FET (red line) and Gr FET (green line) biosensors.
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506.
- Wu, J.T.; Leung, K.; Leung, G.M. Nowcasting and Forecasting the Potential Domestic and International Spread of the 2019-nCoV Outbreak Originating in Wuhan, China: A Modelling Study. Lancet 2020, 395, 689–697.
- Cui, F.; Zhou, H.S. Diagnostic Methods and Potential Portable Biosensors for Coronavirus Disease 2019. Biosens. Bioelectron. 2020, 165, 112349.
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567.
- Tang, Y.-N.; Jiang, D.; Wang, X.; Liu, Y.; Wei, D. Recent Progress on Rapid Diagnosis of COVID-19 by Point-of-Care Testing Platforms. Chin. Chem. Lett. 2023, 108688.
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology 2020, 296, E32–E40.
- Ravi, N.; Cortade, D.L.; Ng, E.; Wang, S.X. Diagnostics for SARS-CoV-2 Detection: A Comprehensive Review of the FDA-EUA COVID-19 Testing Landscape. Biosens. Bioelectron. 2020, 165, 112454.
- Giovannini, G.; Haick, H.; Garoli, D. Detecting COVID-19 from Breath: A Game Changer for a Big Challenge. ACS Sens. 2021, 6, 1408–1417.
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020, 296, E115–E117.
- Etienne, E.E.; Nunna, B.B.; Talukder, N.; Wang, Y.; Lee, E.S. COVID-19 Biomarkers and Advanced Sensing Technologies for Point-of-Care (POC) Diagnosis. Bioengineering 2021, 8, 98.
- Lin, Q.; Wen, D.; Wu, J.; Liu, L.; Wu, W.; Fang, X.; Kong, J. Microfluidic Immunoassays for Sensitive and Simultaneous Detection of IgG/IgM/Antigen of SARS-CoV-2 within 15 Min. Anal. Chem. 2020, 92, 9454–9458.
- Yakoh, A.; Pimpitak, U.; Rengpipat, S.; Hirankarn, N.; Chailapakul, O.; Chaiyo, S. Paper-Based Electrochemical Biosensor for Diagnosing COVID-19: Detection of SARS-CoV-2 Antibodies and Antigen. Biosens. Bioelectron. 2021, 176, 112912.
- Novodchuk, I.; Kayaharman, M.; Prassas, I.; Soosaipillai, A.; Karimi, R.; Goldthorpe, I.A.; Abdel-Rahman, E.; Sanderson, J.; Diamandis, E.P.; Bajcsy, M.; et al. Electronic Field Effect Detection of SARS-CoV-2 N-Protein before the Onset of Symptoms. Biosens. Bioelectron. 2022, 210, 114331.
- Lim, W.Y.; Lan, B.L.; Ramakrishnan, N. Emerging Biosensors to Detect Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): A Review. Biosensors 2021, 11, 434.
- Asif, M.; Ajmal, M.; Ashraf, G.; Muhammad, N.; Aziz, A.; Iftikhar, T.; Wang, J.; Liu, H. The Role of Biosensors in Coronavirus Disease-2019 Outbreak. Curr. Opin. Electrochem. 2020, 23, 174–184.
- Han, S.; Chen, C.; Chen, C.; Wu, L.; Wu, X.; Lu, C.; Zhang, X.; Chao, P.; Lv, X.; Jia, Z.; et al. Coupling Annealed Silver Nanoparticles with a Porous Silicon Bragg Mirror SERS Substrate and Machine Learning for Rapid Non-Invasive Disease Diagnosis. Anal. Chim. Acta 2023, 1254, 341116.
- Dai, C.; Yang, Y.; Xiong, H.; Wang, X.; Gou, J.; Li, P.; Wu, Y.; Chen, Y.; Kong, D.; Yang, Y.; et al. Accurately Detecting Trace-Level Infectious Agents by an Electro-Enhanced Graphene Transistor. Adv. Funct. Mater. 2023, 33, 2300151.
- Fan, Z.; Geng, Z.; Fang, W.; Lv, X.; Su, Y.; Wang, S.; Chen, H. Smartphone Biosensor System with Multi-Testing Unit Based on Localized Surface Plasmon Resonance Integrated with Microfluidics Chip. Sensors 2020, 20, 446.
- Sun, D.; Wu, Y.; Chang, S.-J.; Chen, C.-J.; Liu, J.-T. Investigation of the Recognition Interaction between Glycated Hemoglobin and Its Aptamer by Using Surface Plasmon Resonance. Talanta 2021, 222, 121466.
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Lee, S.Y.; Lertanantawong, B. Rapid Electrochemical Detection of Coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 802.
- Shahdeo, D.; Chauhan, N.; Majumdar, A.; Ghosh, A.; Gandhi, S. Graphene-Based Field-Effect Transistor for Ultrasensitive Immunosensing of SARS-CoV-2 Spike S1 Antigen. ACS Appl. Bio Mater. 2022, 5, 3563–3572.
- Sadighbayan, D.; Hasanzadeh, M.; Ghafar-Zadeh, E. Biosensing Based on Field-Effect Transistors (FET): Recent Progress and Challenges. TrAC Trends Anal. Chem. 2020, 133, 116067.
- Fernandes, R.S.; De Oliveira Silva, J.; Gomes, K.B.; Azevedo, R.B.; Townsend, D.M.; De Paula Sabino, A.; Branco De Barros, A.L. Recent Advances in Point of Care Testing for COVID-19 Detection. Biomed. Pharmacother. 2022, 153, 113538.
- Shabani, E.; Dowlatshahi, S.; Abdekhodaie, M.J. Laboratory Detection Methods for the Human Coronaviruses. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 225–246.
- Kang, H.; Wang, X.; Guo, M.; Dai, C.; Chen, R.; Yang, L.; Wu, Y.; Ying, T.; Zhu, Z.; Wei, D.; et al. Ultrasensitive Detection of SARS-CoV-2 Antibody by Graphene Field-Effect Transistors. Nano Lett. 2021, 21, 7897–7904.
- Cui, T.-R.; Qiao, Y.-C.; Gao, J.-W.; Wang, C.-H.; Zhang, Y.; Han, L.; Yang, Y.; Ren, T.-L. Ultrasensitive Detection of COVID-19 Causative Virus (SARS-CoV-2) Spike Protein Using Laser Induced Graphene Field-Effect Transistor. Molecules 2021, 26, 6947.
- Ji, D.; Zhao, J.; Liu, Y.; Wei, D. Electrical Nanobiosensors for Nucleic Acid Based Diagnostics. J. Phys. Chem. Lett. 2023, 14, 4084–4095.
- Kong, D.; Wang, X.; Gu, C.; Guo, M.; Wang, Y.; Ai, Z.; Zhang, S.; Chen, Y.; Liu, W.; Wu, Y.; et al. Direct SARS-CoV-2 Nucleic Acid Detection by Y-Shaped DNA Dual-Probe Transistor Assay. J. Am. Chem. Soc. 2021, 143, 17004–17014.
- Wu, Y.; Ji, D.; Dai, C.; Kong, D.; Chen, Y.; Wang, L.; Guo, M.; Liu, Y.; Wei, D. Triple-Probe DNA Framework-Based Transistor for SARS-CoV-2 10-in-1 Pooled Testing. Nano Lett. 2022, 22, 3307–3316.
- Ke, G.; Su, D.; Li, Y.; Zhao, Y.; Wang, H.; Liu, W.; Li, M.; Yang, Z.; Xiao, F.; Yuan, Y.; et al. An Accurate, High-Speed, Portable Bifunctional Electrical Detector for COVID-19. Sci. China Mater. 2021, 64, 739–747.
- Hwang, M.T.; Park, I.; Heiranian, M.; Taqieddin, A.; You, S.; Faramarzi, V.; Pak, A.A.; Zande, A.M.; Aluru, N.R.; Bashir, R. Ultrasensitive Detection of Dopamine, IL-6 and SARS-CoV-2 Proteins on Crumpled Graphene FET Biosensor. Adv. Mater. Technol. 2021, 6, 2100712.
- Hwang, M.T.; Heiranian, M.; Kim, Y.; You, S.; Leem, J.; Taqieddin, A.; Faramarzi, V.; Jing, Y.; Park, I.; Van Der Zande, A.M.; et al. Ultrasensitive Detection of Nucleic Acids Using Deformed Graphene Channel Field Effect Biosensors. Nat. Commun. 2020, 11, 1543.
- Gao, J.; Wang, C.; Chu, Y.; Han, Y.; Gao, Y.; Wang, Y.; Wang, C.; Liu, H.; Han, L.; Zhang, Y. Graphene Oxide-Graphene Van Der Waals Heterostructure Transistor Biosensor for SARS-CoV-2 Protein Detection. Talanta 2022, 240, 123197.
More
Information
Subjects:
Chemistry, Analytical
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.8K
Entry Collection:
COVID-19
Revisions:
2 times
(View History)
Update Date:
06 Nov 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




