Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hilda Dinah Kyomuhimbo | -- | 2370 | 2023-09-13 14:44:53 | | | |
| 2 | Fanny Huang | Meta information modification | 2370 | 2023-09-14 08:38:08 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kyomuhimbo, H.D.; Feleni, U.; Haneklaus, N.H.; Brink, H. Application of Enzyme-Nanoparticle-Polymer Composites in Wastewater Treatment. Encyclopedia. Available online: https://encyclopedia.pub/entry/49115 (accessed on 07 February 2026).
Kyomuhimbo HD, Feleni U, Haneklaus NH, Brink H. Application of Enzyme-Nanoparticle-Polymer Composites in Wastewater Treatment. Encyclopedia. Available at: https://encyclopedia.pub/entry/49115. Accessed February 07, 2026.
Kyomuhimbo, Hilda Dinah, Usisipho Feleni, Nils H. Haneklaus, Hendrik Brink. "Application of Enzyme-Nanoparticle-Polymer Composites in Wastewater Treatment" Encyclopedia, https://encyclopedia.pub/entry/49115 (accessed February 07, 2026).
Kyomuhimbo, H.D., Feleni, U., Haneklaus, N.H., & Brink, H. (2023, September 13). Application of Enzyme-Nanoparticle-Polymer Composites in Wastewater Treatment. In Encyclopedia. https://encyclopedia.pub/entry/49115
Kyomuhimbo, Hilda Dinah, et al. "Application of Enzyme-Nanoparticle-Polymer Composites in Wastewater Treatment." Encyclopedia. Web. 13 September, 2023.
Copy Citation
Different water treatment technologies such as photochemical degradation, biodegradation, electrochemical degradation, reverse osmosis, and membrane separation have been used to get rid of water pollutants. Enzymatic treatments have received great attention due to several advantages compared to physical and chemical treatments, such as mild operating conditions and high catalytic efficiency without harsh side effects. Oxidase and peroxidase enzymes from different sources have been immobilized on metal and metal oxide-polymer composites and used in the degradation of pollutants.
enzyme-nanoparticle-polymer composites
wastewater
pollutants
1. Introduction
In recent decades, the global community has increasingly recognized the formidable challenge posed by water pollution arising from the unregulated release of municipal and industrial waste [1][2]. Many industries including petrochemical, paints and explosives, food, pharmaceutical, leather and textile, pulp and paper, and cosmetics have contributed to this cause [3][4]. These discharges cause serious problems to aquatic life due to their high biochemical oxygen demand (BOD), chemical oxygen demand, and blockage of sunlight [5][6].
One of the industries producing the highest level of toxic chemicals from dyeing, printing, and finishing is the leather and textile industry [1]. The conversion of skin into leather in textile industries generates huge amounts of wastewater containing a variety of organic and inorganic chemicals such as dyes, neutral salts, phenols, and biogenic matter of skins [7][8]. The complex aromatic structures of these chemicals, especially the dyes, make them highly soluble in water and stable against light, aerobic decomposition, and oxidizing reagents [9]. Therefore, their accumulation leads to serious environmental concerns for aquatic life and human beings due to their adverse effects of toxicity, carcinogenicity, and mutagenicity [10]. Another industrial sector that has developed rapidly in the last century is the pesticide industry, as it is an important component of modern global agricultural systems for controlling pests and increasing crop yield [11]. These pesticides are applied in much higher doses than those required to kill the pests, and end up accumulating in water bodies via run off and percolation [12]. Unfortunately, these agrochemical residues not only pollute the aquatic systems and damage biodiversity, they cause serious health hazards to humans and may even directly or indirectly lead to death [13][14]. Moreover, these compounds have very long half-lives and can remain in the environment for several decades [15][16].
The growth of the pharmaceutical industry (veterinary and human medicines) in the past years has also led to rising amounts of drugs, antibiotics, and hormones. These medicines are not fully metabolized by living organisms and when these end up in wastewater treatment plants, they are difficult to biodegrade, since most of them are fat soluble [17][18][19]. For example a study conducted by Joss et al. [20] indicated that biological degradation of pharmaceuticals using activated sewage sludge from municipal wastewater could only degrade 4 out of 35 compounds by over 90% and 17 compounds by less than 50%. These compounds have increased in the environment due to their increased consumption and direct discharge into the environment. The presence of pharmaceuticals, cosmetics, and their metabolites in municipal waste and industrial effluents presents a significant challenge, as these compounds cannot be effectively eliminated using conventional techniques, and consequently are released to the receiving environment [21][22]. While in the environment, they accumulate or transform into metabolites under certain environmental conditions, and these secondary metabolites may even be more toxic than the parent compounds [12][23]. These make pathogenic organisms develop resistance against them over time, which is a high risk to human health [24].
The continued release, spread, and accumulation of persistent organic pollutants in the water environment from these industries, including polychlorinated biphenyls and polycyclic aromatic hydrocarbons from the petrochemical industries, have become a major threat to human health due to their toxic, mutagenic, and carcinogenic properties [25][26][27]. The emission of these pollutants occurs at the manufacturing stage, after consumption and disposal of unused products. These products are hard to be tracked or controlled in most situations and are resistant to natural biodegradation [12][28]. Most of these compounds are phenolic and, therefore, bio-recalcitrant, carcinogenic, and easily accumulate in plants and animals. They should, therefore, be removed prior to wastewater discharge [17][29][30].
Different water treatment technologies such as photochemical degradation, biodegradation, electrochemical degradation, reverse osmosis, and membrane separation have been used to get rid of these pollutants. However, these techniques are costly, consist of complicated procedures, do not entirely remove the pollutants, and lead to secondary contaminants that also need to be redisposed of [31][32]. Enzymatic treatments of these pollutants have received great attention due to several advantages compared to physical and chemical treatments, such as mild operating conditions and high catalytic efficiency without harsh side effects [33][34]. Hence, the use of biocatalysts in wastewater treatment has gained momentum due to their ability to target a wide range of pollutants [35]. Enzymes immobilized onto supports are often used in the treatment of wastewaters to ensure improved thermal and pH stability and repeatability, which is rarely achieved with free enzymes [36]. Various pollutants including drugs, dyes, pesticides, polycyclic aromatic hydrocarbons (PAHs), and even heavy metals have been degraded using enzyme/metal-polymer biocatalysts, as demonstrated in Figure 1. Oxidase and peroxidase enzymes from different sources have been immobilized on metal and metal oxide-polymer composites and used in the degradation of pollutants, as observed in Figure 1.
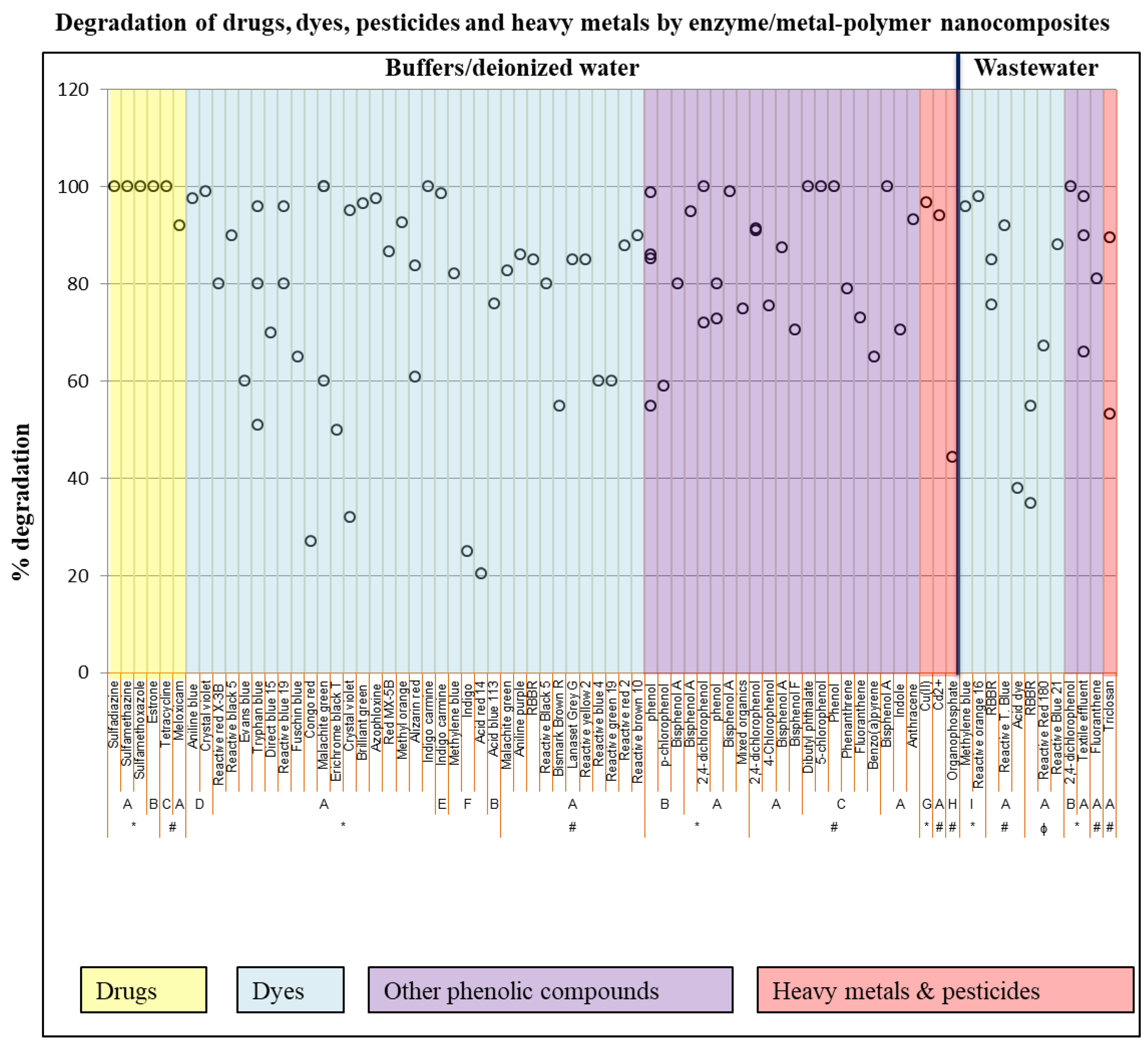
Figure 1. Different pollutants that have been degraded by enzyme-nanoparticle-polymer composites. A—Laccase, B—Horse radish peroxidase, C—Lignin peroxidase, D—Chloroperoxidase, E—Glucose oxidase, F—Glucose oxidase/laccase, G—S. cerevisiae enzyme, H—Glycerophosphodiesterase, I—Manganese peroxidase, * 0–6 h, # 6–24 h, ɸ over 24 h.
2. Laccase-Based Nanocomposite Biocatalysts for Degradation of Pollutants
Laccase is the most explored enzyme in wastewater treatment due to its ability to degrade a wide range of micro pollutants including dyes, pharmaceuticals, and endocrine-disrupting chemicals [37][38][39]. Unlike other oxidoreductases, laccase does not require hydrogen peroxide or other cofactors for substrate cleavage [40][41][42] and its range of compounds for oxidation can be increased with redox mediators [43][44]. Laccase-based composite biocatalysts show great potential in wastewater treatment as they have demonstrated high pollutant degradation rates with high reusability (Table 1). For example, Laccase/Fe2O3/PEI biocatalyst completely degraded sulfa drugs (Sulfadiazine, Sulfamethazine and Sulfamethoxazole) within 30 min and could still degrade 82.8% after 10 cycles in the same time frame [24]. Laccase/Ca-alginate beads degraded 99% bisphenol A [19] and dyes (aniline purple–86%, lanset grey G–85%, and reactive black 5–80%) [45] in 2 h and 24 h, respectively.
Table 1. Application of enzyme-nanoparticle-polymer composites in degradation of organic pollutants for application in wastewater treatment.
| Nanocomposite (NC) | Immobilization Method | Pollutants Removed | Degradation (%) | Degradation Time | Reusability | Ref. |
|---|---|---|---|---|---|---|
| TiO2/polyvinylidene fluoride (PVDF) | Crosslinking of TiO2/PVDF membrane using APTES and glutaraldehyde followed by immersion in laccase solution | Bisphenol A | 95 | 5 h | 91.7% (96 h of continuous use) | [46] |
| TiO2/bacterial cellulose (BC) | Physical adsorption of TiO2 on BC followed by crosslinking with glutaraldehyde and immersion in laccase solution | Reactive red X-3B in presence of ABTS | 80 | 60 min | 70% and 57% (6 and 10 cycles, respectively) | [1] |
| Calcium alginate | Physical entrapment of enzyme in nanocomposite | Fluoranthene in a fluidized bed reactor | 81.06 | 8 h | 66.845% (60 days of storage) | [27] |
| Fe2O3/poly(ethylene glycol)/concovalin A | Chemical co-precipitation followed by crosslinking with glutaraldehyde and immersion in laccase solution | Sulfadiazine | 100 | 30 min | 82.8% (10 consecutive cycles) | [24] |
| Sulfamethazine | ||||||
| Sulfamethoxazole (all in presence of syringaldehyde mediator) |
||||||
| MNPs/chitosan | Physical mixing of NPs and chitosan followed by crosslinking with glutaraldehyde and immersion in laccase solution | Reactive black 5 | 90 | 30 min | 47% (10 cycles) | [47] |
| Evans blue | 60 | 30 min | ||||
| Tryphan blue | 80 | 40 min | ||||
| Direct blue 15 | 70 | 60 min | ||||
| MNPs/polydopamine | Functionalized MNP-polydopamine NC with dialdehyde starch followed by immersion in laccase solution | 2,4-dichlorophenol | 72 | 3 h | 77% (8 cycles) | [48] |
| 91 | 12 h | |||||
| Fe2O3/Cu-alginate | Physical entrapment of enzyme in nanocomposite | Triclosan | 89.6 | 8 h | 86.9% (3 cycles in acetate buffer) | [4] |
| 53.2 | 8 h (wastewater) | |||||
| Remazol Brilliant Blue R (RBBR) | 75.8 | 8 h | ||||
| 55 | 25 h (wastewater) | |||||
| 35 | 25 h (waste water) | |||||
| Cu (II)-chitosan-graft-poly (glycidyl methacrylate)/poly (ethylene imine) | Physical adsorption of laccase on nanocomposites | Phenol in presence of ABTS | 80 | 4 h | 50% (8 cycles) | [30] |
| MNPs/chitosan | Crosslinking with glutaraldehyde followed by immersion in laccase solution | 2,4-Dichlorophenol | 91.4 | 12 h | 75.8% and 57.4% (2,4-DCP and 4-CP after 10 cycles) | [33] |
| 4-Chlorophenol | 75.5 | |||||
| MNPs/SiO2/poly (glycidyl methacrylate)-S-SH | Physical adsorption of enzyme on the nanocomposite | Meloxicam | 92 | 48 h | 82.3%, 88.9%, and 87.5% (meloxicam, piroxicam and Cd2+, respectively, after 5 cycles) | [21] |
| Piroxicam | 95 | |||||
| Cd2+ | 94 | |||||
| MNPs/Poly(p-Phenylenediamine) | Covalent immobilization using glutaraldehyde for crosslinking | Reactive blue 19 | 80 | 1 h | 43% (8 cycles) | [6] |
| MNPs@MoS2/polyethyleneimine | Physical adsorption of laccase on nanocomposite | Malachite green | 82.7 | Overnight | 62% (10 cycles) | [25] |
| Bisphenol A | 87.6 | |||||
| Bisphenol F (all in presence of ABTS) |
70.6 | |||||
| Cu-alginate | Physical entrapment of enzyme in nanocomposite | Fuschin blue | 65 (HOBT) | 4 h | 100% and 95% (120 h continuous use and 15 days storage, respectively) | [8] |
| Congo red | 27 (ABTS) | |||||
| Tryphan blue | 51(syringaldehyde) | |||||
| Malachite green | 60 (ABTS) | |||||
| Erichrome black T | 50 (HOBT) | |||||
| Crystal violet (all in different mediators) |
32 (HOBT) | |||||
| Textile effluent in a continuous flow packed bed bioreactor | 66 (colour) 90 (BOD) 98 (COD) |
|||||
| MNPs/chitosan | Physical entrapment of enzyme in presence of ionic liquid and ABTS | 2,4-dichlorophenol | 100 | 4 h | 93.2% (for 2,4-DCP after 6 cycles) | [49] |
| Bisphenol A | 100 | 72 h | ||||
| Indole | 70.5 | 72 h | ||||
| Anthracene | 93.3 | 72 h | ||||
| MNPs/polyethylenimine | Crosslinking of NPs with PEI using glutaraldehyde followed by chelation of laccase with Cu(II) | Phenol in a fixed bed reactor | 72.93% at a flowrate of 25 μL/min | - | - | [34] |
| MNPs/Cu2+-PEG | In situ oxidation of metal salt using PEG followed by physical adsorption of laccase | Malachite green | 100 (ABTS) | 120 min | 99.9, 90.1, 89.4, 94.6, 76.5, 80.1, 74.6, and 66.1% (respectively, for the dyes after 10 cycles) | [10] |
| Brilliant green | 96.5 (ABTS) | |||||
| Crystal violet | 95.2 (ABTS) | |||||
| Azophloxine | 97.7 (TEMPO) | |||||
| Red MX-5B | 86.6 (ABTS) | |||||
| Methyl orange | 92.7 (VLA) | |||||
| Reactive blue 19 | 96 (TEMPO) | |||||
| Alizarin red | 83.7 (TEMPO) | |||||
| TiO2/Zn-alginate | Physical entrapment of enzyme in nanocomposite | Alizarin red | 61 | 5 h | 100% (14 cycles) | [50] |
| Tryphan blue | 96 | |||||
| Malachite green | 100 | |||||
| Indigo carmine | 100 | |||||
| Ca-alginate | Physical entrapment with crosslinking of enzyme prior to entrapment | Bisphenol A | 99 | 2 h | 70% (10 successive cycles) | [19] |
| Ca-alginate | Physical entrapment of enzyme in nanocomposite | Aniline purple | 86.1 | 24 h | - | [51] |
| Ca-alginate | Physical entrapment of enzyme in nanocomposite | Reactive Red 180 | 67.2 | 11 days | - | [52] |
| Reactive Blue 21 | 88.05 | |||||
| Ca-alginate | Physical entrapment of enzyme in nanocomposite | Reactive T. Blue | 92 | 72 h | 22.3% (6 cycles) | [53] |
| Ca-alginate | Physical entrapment of enzyme in nanocomposite | RBBR | 85 | 2 h | 52.1% and 70% (Bismarck brown and all the others, respectively) | [45] |
| Reactive Black 5 | 80 | 24 h | ||||
| Bismarck Brown R | 55 | 24 h | ||||
| Lancet Grey G | 85 | 24 h | ||||
| Cu-alginate | Physical entrapment of enzyme in nanocomposite | Acid dye | 38% | 24 h | - | [54] |
| MNPs/chitosan | Crosslinking with glutaraldehyde followed by adsorption in laccase solution | Reactive yellow 2 | 85 | 10 h | - | [55] |
| Reactive blue 4 | 60 | 12 h | ||||
| MNPs/poly(GMA-MMA)/Cu-Poly(4-vinyl pyridine | Polymer grafting with Cu chelation followed by adsorption of enzyme | Reactive green 19 | 60 | 18 h | 63%, 76%, and 59% (green, red, and brown dyes, respectively) | [56] |
| Reactive red 2 | 88 | |||||
| Reactive brown 10 | 90 | |||||
| Cu-alginate | Physical entrapment of enzyme in nanocomposite | phenol model solution containing tannic acid, gallic acid, ferulic acid, resorcinol, and pyrogallol | 75 | 6 h | 35% (8 cycles) | [57] |
| FScubes/PDA@PVDF | Prepared the FS/PDA@PVDF membrane using solvothermal process followed by covalent immobilization of laccase using glutaraldehyde as cross linker | Congo red | 97.1 | 3 h | 85% and 76% (7 days and 5 cycles, respectively) | [58] |
3. Horse Radish Peroxidase (HRP)-Based Nanocomposite Biocatalysts for Degradation of Pollutants
Another commonly explored peroxidase on nanoparticle-polymer composite materials is horse radish peroxidase (HRP), due to its ability to oxidize a wide range of phenolic compounds in the presence of hydrogen peroxide [59]. It oxidizes phenolic compounds by adding hydrogen peroxide to form corresponding radicals which spontaneously interact to form insoluble polymers that can be easily removed from the wastewater [60]. HRP/nanoparticle-polymer composite biocatalysts have been explored in the degradation of phenols, dyes, and endocrine-disrupting compounds, as illustrated in Table 2. For example, HRP/MNPs/polyvinyl alcohol/poly acrylic acid could completely degrade estrone after 40 min [18], and HRP/TiO2/polydopamine completely removed 2,4-dicholorphenol in Zhaohe wastewater samples in only 30 min [61]. Interestingly, the HRP/TiO2/polydopamine biocatalyst retained 100% and 90% degradation activity after 15 and 25 reuses, respectively.
Table 2. Application of enzyme-nanoparticle-polymer composites in degradation of organic pollutants for application in wastewater treatment.
| Nanocomposite (NC) | Immobilization Method | Pollutants Removed | Degradation (%) | Degradation Time | Reusability | Ref. |
|---|---|---|---|---|---|---|
| TiO2/polydopamine | In situ polymerization of dopamine on TiO2NPs followed by covalent crosslinking of enzyme with glutaraldehyde | 2,4-dichlorophenol | 100 | 30 min | 100%, 90%, and 63.6% (15, 25, and 40 reuses, respectively) | [61] |
| MNPs/poly(glycidylmethacrylate-co-methylmethacrylate) (poly(GMA-MMA)) | Crosslinking of enzyme and nanocomposite beads using glutaraldehyde | phenol | 86 | 2 h | 84% (8 weeks), 92%, and 79% (phenol and p-chlorophenol, respectively, after 48 h of continuous use) | [3] |
| p-chlorophenol (in the presence of H2O2) |
59 | |||||
| Fe2O3/poly (amido amine) (PAMAM)/silk fibroin | Crosslinking of enzyme with nanocomposites using glutaraldehyde | Bisphenol A in presence of H2O2 | 80 | 120 min | - | [62] |
| Calcium alginate | Physical entrapment of enzyme in nanocomposite | Acid blue 113 | 76 | 240 min | Can be recycled up to 3 times | [7] |
| Aluminosilicate halloysite nanotubes/chitosan | Crosslinking of enzyme with nanocomposites using glutaraldehyde | Phenol in presence of hydrogen peroxide | 98.8 | 30 min | 60% (4 cycles) | [63] |
| MNPs/polyacrylonitrile | Crosslinking of enzyme with nanocomposites using glutaraldehyde | Phenol | 85.2 | - | 52% (5 cycles) | [29] |
| MNPs/poly(vinyl alcohol)/poly(acrylic acid) | Physical adsorption of enzyme on nanocomposites | Estrone | 100 | 40 min | 56.2% (7 cycles) | [18] |
| MNPs/polymethyl methacrylate | Physical entrapment of enzyme in nanocomposite | Phenol in presence of hydrogen peroxide | 55 | 50 min | - | [64] |
| MNPs/poly(glycidylmethacrylate-co-methylmethacrylate) (poly(GMA-MMA)) | Crosslinking of enzyme with nanocomposite beads using glutaraldehyde | Phenol | 86 | 2 h | 91% and 79% (phenol and chlorophenol, respectively, after 48 h of continuous operation) | [3] |
| p-Chlorophenol (in presence of hydrogen peroxide in a fluidized bed reactor) |
59 |
4. Other Oxidase and Peroxidase-Based Nanocomposite Biocatalysts for Degradation of Pollutants
Other enzymes such as chloroperoxidase, manganese peroxidase, and lignin peroxidase immobilized on composite materials, though not very popular, prove that they can offer wonderful materials for pollutant degradation (Table 3). For example, when lignin peroxidase was immobilized on MNPs@SiO2/polydopamine, it was able to degrade tetracycline and other phenolics such as 5-chlorophenol, phenol, and dibutyl phthalate completely within 24 h [32]. Manganese peroxidase immobilized on MNPs/chitosan degraded 96% of methylene blue in synthetic wastewater in just 50 min [2], glucose oxidase immobilized on NiFe2O4/tannin could degrade 98.6% of indigo carmine in presence of UV light within 90 min [31], and chloroperoxidase/TiO2/polydopamine nanocomposites degraded over 95% of aniline blue and crystal violet in 2 min [61].
Table 3. Application of enzyme-nanoparticle-polymer composites in degradation of organic pollutants for application in wastewater treatment.
| Nanocomposite (NC) | Enzyme | Immobilization Method | Pollutants Removed | Degradation (%) | Degradation Time | Reusability | Ref. |
|---|---|---|---|---|---|---|---|
| iO2/polydopamine | Chloroperoxidase (CPO) | Covalent crosslinking of enzyme with nanocomposites using glutaraldehyde | Aniline blue | 97.58 | 2 min | 90.3%, 78.2%, and 53.71% (10, 15, and 20 reuses, respectively) | [61] |
| Crystal violet | 98.98 | 2 min | |||||
| NiFe2O4/tannin | Glucose oxidase | Physical adsorption of enzyme on nanocomposite | Indigo carmine in presence of UV light | 98.6 | 90 min | 85.57% (5 cycles) | [31] |
| MnFe2O4/calcium alginate | Glucose oxidase and Laccase |
Physical adsorption of enzymes on the nanocomposite | Methylene blue | 82.13 | 1 h | - | [9] |
| Indigo | 25.09 | ||||||
| Acid red 14 | 20.42 | ||||||
| MNPs/PAMAM | Glycerophosphodiesterase (GpdQ) | Crosslinking of enzyme with nanocomposites using glutaraldehyde | Organophosphate pesticide | 44.5 | 120 days | Used as a filter in a Pasteur pipette between two layers of sand | [14] |
| MNPs@SiO2/polydopamine | Lignin peroxidase | Physical adsorption of enzymes on the nanocomposite | Tetracycline | 100 | 24 h | 80.3% and 67.5% (7 and 14 days of storage), 70% and 30% (4 and 8 cycles, respectively) | [32] |
| Dibutyl phthalate | 100 | 24 h | |||||
| 5-chlorophenol | 100 | 24 h | |||||
| Phenol | 100 | 24 h | |||||
| Phenanthrene | 79 | 24 h | |||||
| Fluoranthene | 73 | 24 h | |||||
| Benzo(a)pyrene | 65 | 24 h | |||||
| MNPs/chitosan | Manganese peroxidase | Crosslinking of enzyme with nanocomposites using glutaraldehyde | Methylene blue | 96 | 50 min | 91.7% and 86.7% (5 cycles-methylene blue and reactive orange, respectively) | [2] |
| Reactive orange 16 | 98 | ||||||
| Fe2O3/chitosan | Saccharomyces cerevisiae enzyme | Adsorption of chitosan on the NPs surface followed by crosslinking with enzyme using glutaraldehyde | Cu(II) | 96.8 | 60 min | - | [65] |
References
- Li, G.; Nandgaonkar, A.G.; Wang, Q.; Zhang, J.; Krause, W.E.; Wei, Q.; Lucia, L.A. Laccase-immobilized bacterial cellulose/TiO2 functionalized composite membranes: Evaluation for photo- and bio-catalytic dye degradation. J. Membr. Sci. 2017, 525, 89–98.
- Siddeeg, S.M.; Tahoon, M.A.; Mnif, W.; Ben Rebah, F. Iron Oxide/Chitosan Magnetic Nanocomposite Immobilized Manganese Peroxidase for Decolorization of Textile Wastewater. Processes 2020, 8, 5.
- Bayramoğlu, G.; Arıca, M.Y. Enzymatic removal of phenol and p-chlorophenol in enzyme reactor: Horseradish peroxidase immobilized on magnetic beads. J. Hazard. Mater. 2008, 156, 148–155.
- Le, T.T.; Murugesan, K.; Lee, C.-S.; Vu, C.H.; Chang, Y.-S.; Jeon, J.-R. Degradation of synthetic pollutants in real wastewater using laccase encapsulated in core–shell magnetic copper alginate beads. Bioresour. Technol. 2016, 216, 203–210.
- Lin, C.-W.; Wu, C.-H.; Huang, W.-T.; Tsai, S.-L. Evaluation of different cell-immobilization strategies for simultaneous distillery wastewater treatment and electricity generation in microbial fuel cells. Fuel 2015, 144, 1–8.
- Liu, Y.; Yan, M.; Geng, Y.; Huang, J. Laccase Immobilization on Poly(p-Phenylenediamine)/Fe3O4 Nanocomposite for Reactive Blue 19 Dye Removal. Appl. Sci. 2016, 6, 232.
- Preethi, S.; Anumary, A.; Ashokkumar, M.; Thanikaivelan, P. Probing horseradish peroxidase catalyzed degradation of azo dye from tannery wastewater. SpringerPlus 2013, 2, 341.
- Sondhi, S.; Kaur, R.; Kaur, S.; Kaur, P.S. Immobilization of laccase-ABTS system for the development of a continuous flow packed bed bioreactor for decolorization of textile effluent. Int. J. Biol. Macromol. 2018, 117, 1093–1100.
- Shojaat, R.; Saadatjoo, N.; Karimi, A.; Aber, S. Simultaneous adsorption–degradation of organic dyes using MnFe2O4/calcium alginate nano-composites coupled with GOx and laccase. J. Environ. Chem. Eng. 2016, 4, 1722–1730.
- Li, Z.; Zhu, Q.; Liu, Z.; Sha, L.; Chen, Z. Improved performance of immobilized laccase for catalytic degradation of synthetic dyes using redox mediators. New J. Chem. 2022, 46, 9792–9798.
- Bapat, G.; Labade, C.; Chaudhari, A.; Zinjarde, S. Silica nanoparticle based techniques for extraction, detection, and degradation of pesticides. Adv. Colloid Interface Sci. 2016, 237, 1–14.
- Feng, S.; Hao Ngo, H.; Guo, W.; Woong Chang, S.; Duc Nguyen, D.; Cheng, D.; Varjani, S.; Lei, Z.; Liu, Y. Roles and applications of enzymes for resistant pollutants removal in wastewater treatment. Bioresour. Technol. 2021, 335, 125278.
- Bilal, M.; Iqbal, H.M.N.; Barceló, D. Persistence of pesticides-based contaminants in the environment and their effective degradation using laccase-assisted biocatalytic systems. Sci. Total Environ. 2019, 695, 133896.
- Daumann, L.J.; Larrabee, J.A.; Ollis, D.; Schenk, G.; Gahan, L.R. Immobilization of the enzyme GpdQ on magnetite nanoparticles for organophosphate pesticide bioremediation. J. Inorg. Biochem. 2014, 131, 1–7.
- Bucur, B.; Munteanu, F.-D.; Marty, J.-L.; Vasilescu, A. Advances in Enzyme-Based Biosensors for Pesticide Detection. Biosensors 2018, 8, 27.
- Xiong, S.; Deng, Y.; Zhou, Y.; Gong, D.; Xu, Y.; Yang, L.; Chen, H.; Chen, L.; Song, T.; Luo, A.; et al. Current progress in biosensors for organophosphorus pesticides based on enzyme functionalized nanostructures: A review. Anal. Methods 2018, 10, 5468–5479.
- Zdarta, J.; Jankowska, K.; Bachosz, K.; Degórska, O.; Kaźmierczak, K.; Nguyen, L.N.; Nghiem, L.D.; Jesionowski, T. Enhanced Wastewater Treatment by Immobilized Enzymes. Curr. Pollut. Rep. 2021, 7, 167–179.
- Xu, R.; Yuan, J.; Si, Y.; Li, F.; Zhang, B. Estrone removal by horseradish peroxidase immobilized on a nanofibrous support with Fe 3 O 4 nanoparticles. RSC Adv. 2016, 6, 3927–3933.
- Lassouane, F.; Aït-Amar, H.; Amrani, S.; Rodriguez-Couto, S. A promising laccase immobilization approach for Bisphenol A removal from aqueous solutions. Bioresour. Technol. 2019, 271, 360–367.
- Joss, A.; Zabczynski, S.; Göbel, A.; Hoffmann, B.; Löffler, D.; McArdell, C.S.; Ternes, T.A.; Thomsen, A.; Siegrist, H. Biological degradation of pharmaceuticals in municipal wastewater treatment: Proposing a classification scheme. Water Res. 2006, 40, 1686–1696.
- Siddeeg, S.M.; Amari, A.; Tahoon, M.A.; Alsaiari, N.S.; Rebah, F.B. Removal of meloxicam, piroxicam and Cd+2 by Fe3O4/SiO2/glycidyl methacrylate-S-SH nanocomposite loaded with laccase. Alex. Eng. J. 2020, 59, 905–914.
- Sarkar, S.; Chakraborty, S.; Bhattacharjee, C. Photocatalytic degradation of pharmaceutical wastes by alginate supported TiO2 nanoparticles in packed bed photo reactor (PBPR). Ecotoxicol. Environ. Saf. 2015, 121, 263–270.
- Adewuyi, A. Chemically Modified Biosorbents and Their Role in the Removal of Emerging Pharmaceutical Waste in the Water System. Water 2020, 12, 1551.
- Shi, L.; Ma, F.; Han, Y.; Zhang, X.; Yu, H. Removal of sulfonamide antibiotics by oriented immobilized laccase on Fe3O4 nanoparticles with natural mediators. J. Hazard. Mater. 2014, 279, 203–211.
- Ran, F.; Zou, Y.; Xu, Y.; Liu, X.; Zhang, H. Fe3O4@MoS2@PEI-facilitated enzyme tethering for efficient removal of persistent organic pollutants in water. Chem. Eng. J. 2019, 375, 121947.
- Sharma, B.; Dangi, A.K.; Shukla, P. Contemporary enzyme based technologies for bioremediation: A review. J. Environ. Manag. 2018, 210, 10–22.
- Xie, H.; Chen, Y.; Wang, C.; Shi, W.; Zuo, L.; Xu, H. The removal of fluoranthene by Agaricus bisporus immobilized in Ca-alginate modified by Lentinus edodes nanoparticles. RSC Adv. 2015, 5, 44812–44823.
- Varga, B.; Somogyi, V.; Meiczinger, M.; Kováts, N.; Domokos, E. Enzymatic treatment and subsequent toxicity of organic micropollutants using oxidoreductases—A review. J. Clean. Prod. 2019, 221, 306–322.
- Li, J.; Chen, X.; Xu, D.; Pan, K. Immobilization of horseradish peroxidase on electrospun magnetic nanofibers for phenol removal. Ecotoxicol. Environ. Saf. 2019, 170, 716–721.
- Alver, E.; Metin, A.Ü. Chitosan based metal-chelated copolymer nanoparticles: Laccase immobilization and phenol degradation studies. Int. Biodeterior. Biodegrad. 2017, 125, 235–242.
- Atacan, K.; Güy, N.; Çakar, S.; Özacar, M. Efficiency of glucose oxidase immobilized on tannin modified NiFe2O4 nanoparticles on decolorization of dye in the Fenton and photo-biocatalytic processes. J. Photochem. Photobiol. A Chem. 2019, 382, 111935.
- Guo, J.; Liu, X.; Zhang, X.; Wu, J.; Chai, C.; Ma, D.; Chen, Q.; Xiang, D.; Ge, W. Immobilized lignin peroxidase on Fe3O4@SiO2@polydopamine nanoparticles for degradation of organic pollutants. Int. J. Biol. Macromol. 2019, 138, 433–440.
- Zhang, K.; Yang, W.; Liu, Y.; Zhang, K.; Chen, Y.; Yin, X. Laccase immobilized on chitosan-coated Fe3O4 nanoparticles as reusable biocatalyst for degradation of chlorophenol. J. Mol. Struct. 2020, 1220, 128769.
- Xia, T.-T.; Feng, M.; Liu, C.-L.; Liu, C.-Z.; Guo, C. Efficient phenol degradation by laccase immobilized on functional magnetic nanoparticles in fixed bed reactor under high-gradient magnetic field. Eng. Life Sci. 2021, 21, 374–381.
- Morshed, M.N.; Behary, N.; Bouazizi, N.; Guan, J.; Nierstrasz, V.A. An overview on biocatalysts immobilization on textiles: Preparation, progress and application in wastewater treatment. Chemosphere 2021, 279, 130481.
- Kołodziejczak-Radzimska, A.; Nghiem, L.D.; Jesionowski, T. Functionalized Materials as a Versatile Platform for Enzyme Immobilization in Wastewater Treatment. Curr. Pollut. Rep. 2021, 7, 263–276.
- Jahangiri, E.; Thomas, I.; Schulze, A.; Seiwert, B.; Cabana, H.; Schlosser, D. Characterisation of electron beam irradiation-immobilised laccase for application in wastewater treatment. Sci. Total Environ. 2018, 624, 309–322.
- Bilal, M.; Iqbal, H.M.N.; Barceló, D. Mitigation of bisphenol A using an array of laccase-based robust bio-catalytic cues—A review. Sci. Total Environ. 2019, 689, 160–177.
- Ren, D.; Wang, Z.; Jiang, S.; Yu, H.; Zhang, S.; Zhang, X. Recent environmental applications of and development prospects for immobilized laccase: A review. Biotechnol. Genet. Eng. Rev. 2020, 36, 81–131.
- Unuofin, J.O.; Okoh, A.I.; Nwodo, U.U. Aptitude of Oxidative Enzymes for Treatment of Wastewater Pollutants: A Laccase Perspective. Molecules 2019, 24, 2064.
- Singh, J.; Saharan, V.; Kumar, S.; Gulati, P.; Kapoor, R. Laccase grafted membranes for advanced water filtration systems: A green approach to water purification technology. Crit. Rev. Biotechnol. 2017, 38, 883–901.
- Daronch, N.A.; Kelbert, M.; Pereira, C.S.; de Araújo, P.H.H.; de Oliveira, D. Elucidating the choice for a precise matrix for laccase immobilization: A review. Chem. Eng. J. 2020, 397, 125506.
- Deska, M.; Kończak, B. Immobilized fungal laccase as “green catalyst” for the decolourization process—State of the art. Process Biochem. 2019, 84, 112–123.
- Okwara, P.C.; Afolabi, I.S.; Ahuekwe, E.F. Application of laccase in aflatoxin B1 degradation: A review. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1107, 012178.
- Daâssi, D.; Rodríguez-Couto, S.; Nasri, M.; Mechichi, T. Biodegradation of textile dyes by immobilized laccase from Coriolopsis gallica into Ca-alginate beads. Int. Biodeterior. Biodegrad. 2014, 90, 71–78.
- Hou, J.; Dong, G.; Ye, Y.; Chen, V. Enzymatic degradation of bisphenol-A with immobilized laccase on TiO2 sol–gel coated PVDF membrane. J. Membr. Sci. 2014, 469, 19–30.
- Nadaroglu, H.; Mosber, G.; Gungor, A.A.; Adıguzel, G.; Adiguzel, A. Biodegradation of some azo dyes from wastewater with laccase from Weissella viridescens LB37 immobilized on magnetic chitosan nanoparticles. J. Water Process Eng. 2019, 31, 100866.
- Chen, C.; Sun, W.; Lv, H.; Li, H.; Wang, Y.; Wang, P. Spacer arm-facilitated tethering of laccase on magnetic polydopamine nanoparticles for efficient biocatalytic water treatment. Chem. Eng. J. 2018, 350, 949–959.
- Qiu, X.; Wang, S.; Miao, S.; Suo, H.; Xu, H.; Hu, Y. Co-immobilization of laccase and ABTS onto amino-functionalized ionic liquid-modified magnetic chitosan nanoparticles for pollutants removal. J. Hazard. Mater. 2021, 401, 123353.
- Khakshoor, M.; Makhdoumi, A.; Asoodeh, A.; Hosseindokht, M.R. Co-immobilized spore laccase/TiO2 nanoparticles in the alginate beads enhance dye removal by two-step decolorization. Environ. Sci. Pollut. Res. 2021, 28, 6099–6110.
- Tišma, M.; Šalić, A.; Planinić, M.; Zelić, B.; Potočnik, M.; Šelo, G.; Bucić-Kojić, A. Production, characterisation and immobilization of laccase for an efficient aniline-based dye decolourization. J. Water Process Eng. 2020, 36, 101327.
- Velu, C.; Veeramani, E.; Suntharam, S.; Kalimuthu, K. Insilico Screening and Comparative Study on the Effectiveness of Textile Dye Decolourization by Crude Laccase Immobilised Alginate Encapsulated Beads from Pleurotus ostreatus. J. Bioprocess. Biotech. 2011, 1, 4.
- Noreen, S.; Asgher, M.; Hussain, F.; Iqbal, A. Performance Improvement of Ca-Alginate Bead Cross- Linked Laccase from Trametes versicolor IBL-04. BioResources 2016, 11, 558–572.
- Faraco, V.; Pezzella, C.; Miele, A.; Giardina, P.; Sannia, G. Bio-remediation of colored industrial wastewaters by the white-rot fungi Phanerochaete chrysosporium and Pleurotus ostreatus and their enzymes. Biodegradation 2009, 20, 209–220.
- Bayramoglu, G.; Yilmaz, M.; Yakup Arica, M. Preparation and characterization of epoxy-functionalized magnetic chitosan beads: Laccase immobilized for degradation of reactive dyes. Bioprocess Biosyst. Eng. 2010, 33, 439–448.
- Bayramoğlu, G.; Yilmaz, M.; Yakup Arica, M. Reversible immobilization of laccase to poly(4-vinylpyridine) grafted and Cu(II) chelated magnetic beads: Biodegradation of reactive dyes. Bioresour. Technol. 2010, 101, 6615–6621.
- Niladevi, K.N.; Prema, P. Immobilization of laccase from Streptomyces psammoticus and its application in phenol removal using packed bed reactor. World J. Microbiol. Biotechnol. 2008, 24, 1215–1222.
- Zhu, Y.; Qiu, F.; Rong, J.; Zhang, T.; Mao, K.; Yang, D. Covalent laccase immobilization on the surface of poly(vinylidene fluoride) polymer membrane for enhanced biocatalytic removal of dyes pollutants from aqueous environment. Colloids Surf. B Biointerfaces 2020, 191, 111025.
- Kumar, V.; Misra, N.; Kumar Goel, N.; Thakar, R.; Gupta, J.; Varshney, L. A horseradish peroxidase immobilized radiation grafted polymer matrix: A biocatalytic system for dye waste water treatment. RSC Adv. 2016, 6, 2974–2981.
- Wang, S.; Fang, H.; Wen, Y.; Cai, M.; Liu, W.; He, S.; Xu, X. Applications of HRP-immobilized catalytic beads to the removal of 2,4-dichlorophenol from wastewater. RSC Adv. 2015, 5, 57286–57292.
- Cheng, H.; Hu, M.; Zhai, Q.; Li, S.; Jiang, Y. Polydopamine tethered CPO/HRP-TiO2 nano-composites with high bio-catalytic activity, stability and reusability: Enzyme-photo bifunctional synergistic catalysis in water treatment. Chem. Eng. J. 2018, 347, 703–710.
- Xu, J.; Tang, T.; Zhang, K.; Ai, S.; Du, H. Electroenzymatic catalyzed oxidation of bisphenol-A using HRP immobilized on magnetic silk fibroin nanoparticles. Process Biochem. 2011, 46, 1160–1165.
- Zhai, R.; Zhang, B.; Wan, Y.; Li, C.; Wang, J.; Liu, J. Chitosan–halloysite hybrid-nanotubes: Horseradish peroxidase immobilization and applications in phenol removal. Chem. Eng. J. 2013, 214, 304–309.
- Abdulaal, W.H.; Almulaiky, Y.Q.; El-Shishtawy, R.M. Encapsulation of HRP Enzyme onto a Magnetic Fe3O4 Np–PMMA Film via Casting with Sustainable Biocatalytic Activity. Catalysts 2020, 10, 181.
- Peng, Q.; Liu, Y.; Zeng, G.; Xu, W.; Yang, C.; Zhang, J. Biosorption of copper(II) by immobilizing Saccharomyces cerevisiae on the surface of chitosan-coated magnetic nanoparticles from aqueous solution. J. Hazard. Mater. 2010, 177, 676–682.
More
Information
Subjects:
Polymer Science
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Entry Collection:
Wastewater Treatment
Revisions:
2 times
(View History)
Update Date:
14 Sep 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




