Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Sun, J.; Yao, K.; Huang, G.; Zhang, C.; Leach, M.; Huang, K.; Yang, X. Skin Lesion Datasets and Image Preprocessing. Encyclopedia. Available online: https://encyclopedia.pub/entry/42892 (accessed on 04 March 2026).
Sun J, Yao K, Huang G, Zhang C, Leach M, Huang K, et al. Skin Lesion Datasets and Image Preprocessing. Encyclopedia. Available at: https://encyclopedia.pub/entry/42892. Accessed March 04, 2026.
Sun, Jie, Kai Yao, Guangyao Huang, Chengrui Zhang, Mark Leach, Kaizhu Huang, Xi Yang. "Skin Lesion Datasets and Image Preprocessing" Encyclopedia, https://encyclopedia.pub/entry/42892 (accessed March 04, 2026).
Sun, J., Yao, K., Huang, G., Zhang, C., Leach, M., Huang, K., & Yang, X. (2023, April 10). Skin Lesion Datasets and Image Preprocessing. In Encyclopedia. https://encyclopedia.pub/entry/42892
Sun, Jie, et al. "Skin Lesion Datasets and Image Preprocessing." Encyclopedia. Web. 10 April, 2023.
Copy Citation
Skin lesions affect millions of people worldwide. They can be easily recognized based on their typically abnormal texture and color but are difficult to diagnose due to similar symptoms among certain types of lesions.
skin image segmentation
skin lesion classification
machine learning
deep learning
1. Introduction
The skin protects the human body from outside hazardous substances and is the largest organ in the human body. Skin diseases can be induced by various causes, such as fungal infections, bacterial infections, allergies, or viruses [1]. Skin lesions, as one of the most common diseases, have affected a large population worldwide.
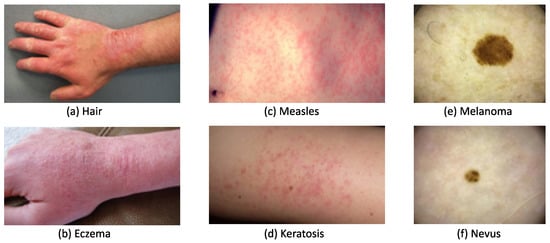
A skin lesion can be easily recognized based on its abnormal texture and/or color, but it can be difficult to accurately diagnose due to similar symptoms occurring with different types of lesions. Figure 1a,b, show that both contact dermatitis and eczema present similarly with redness, swelling, and chapping on a hand. Similarly, as seen in Figure 1c,d, measles and keratosis show red dots that are sporadically distributed on the skin. These similarities lower diagnosis accuracy, particularly for dermatologists with less than two years of clinical experience. To administer appropriate treatment, it is essential to identify skin lesions correctly and as early as possible. Early diagnosis usually leads to a better prognosis and increases the chance of full recovery in most instances. The training of a dermatologist requires several years of clinical experience as well as high education costs. The average tuition fee in medical school was USD 218,792 in 2021 has risen to around USD 1500 each year since 2013 [2]. An automated solution for skin disease diagnosis is becoming increasingly urgent. This is especially true in developing countries, which often lack the advanced medical equipment and expertise
Artificial intelligence (AI) has made rapid progress in image recognition over the last decade, using a combination of methods such as machine learning (ML) and deep learning (DL). Segmentation and classification models can be developed using traditional ML and DL methods. Traditional ML methods have simpler structures compared to DL methods, which use extracted and selected features as inputs [4]. The DL method can automatically discover underlying patterns and identify the most descriptive and salient features in image recognition and processing tasks. This progress has significantly motivated medical workers to explore the potential for the application of AI methods in disease diagnosis, particularly for skin disease diagnosis. Moreover, DL has already demonstrated its capability in this field by achieving similar diagnostic accuracy as dermatologists with 5 years of clinical experience [5]. Therefore, DL is considered a potential tool for cost-effective skin health diagnosis.
The success of AI applications heavily relies on the support of big data to ensure reliable performance and generalization ability. To speed up the progress of AI application in skin disease diagnosis, it is essential to establish reliable databases. Researchers from multiple organizations worked together to create the International Skin Imaging Collaboration (ISIC) Dataset for skin disease research and organized challenges from 2016 to 2020 [6]. The most common research task is to detect melanoma, a life-threatening skin cancer. According to the World Health Organization (WHO), the number of melanoma cases is expected to increase to 466,914 with associated deaths increasing to 105,904 by 2040 [7].
Several additional datasets with macroscopic or dermoscopic images are publicly accessible on the internet, such as the PH2 dataset [8], the Human Against Machine with 10,000 training images (HAM 10,000) dataset [9], the BCN 20,000 dataset [10], the Interactive Atlas of Dermoscopy (EDRA) dataset [11], and the Med-Node dataset [12]. Such datasets can be used individually, partially, or combined based on the specific tasks under investigation.
Macroscopic or close-up images of skin lesions on the human body are often taken using common digital cameras or smartphone cameras. Dermoscopic images, on the other hand, are collected using a standard procedure in preliminary diagnosis and produce images containing fewer artifacts and more detailed features, which can visualize the skin’s inner layer (invisible to the naked eye). Although images may vary with shooting distance, illumination conditions, and camera resolution, dermoscopic images are more likely to achieve higher performance in segmentation and classification than macroscopic images [13].
To conduct a rigorous and thorough review of skin lesion detection studies, researchers utilized a systematic search strategy that involved the PubMed database—an authoritative, widely-used resource for accessing biomedical and life sciences literature. The analysis primarily focused on studies published after 2013 that explored the potential applications of ML in skin lesion diagnosis. To facilitate this analysis, researchers sourced high-quality datasets from reputable educational and medical organizations. These datasets served as valuable tools not only for developing and testing new diagnostic algorithms but also for educating the broader public on skin health and the importance of early detection. By utilizing these datasets in the research, researchers gained a deeper understanding of the current state-of-the-art in skin lesion detection and identified areas for future research and innovation.
Researchers discuss the most relevant skin recognition studies, including datasets, image preprocessing, up-to-date traditional ML and DL applications in segmentation and classification, and challenges based on the original research or review papers from journals and scientific conferences in ScienceDirect, IEEE, and SpringerLink databases. The common schematic diagram of the automated skin image diagnosis procedure is shown in Figure 2.
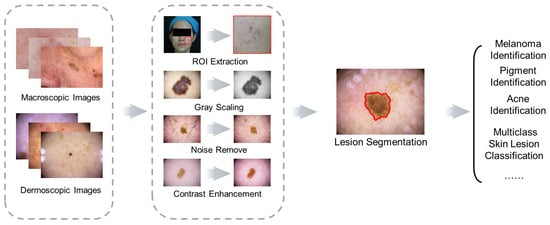
Figure 2. Schematic diagram of skin image diagnosis.
2. Skin Lesion Datasets and Image Preprocessing
2.1. Skin Lesion Datasets
To develop skin diagnosis models, various datasets of different sizes and skin lesion types have been created by educational institutions and medical organizations. These datasets can serve as platforms to educate the general public and as tools to test newly developed diagnosis algorithms.
The first dermoscopic image dataset, PH2, had 200 carefully selected images with segmented lesions, as well as clinical and histological diagnosis records. The Med-Node dataset contains a total of 170 macroscopic images, including 70 melanoma and 100 nevus images, collected by the Department of Dermatology, University Medical Center Groningen. Due to the limited image quantity and classes, these labeled datasets are often used together with others in the development of diagnosis models.
The most popular skin disease detection dataset, ISIC Archive, was created through the collaboration of 30 academic centers and companies worldwide with the aim of improving early melanoma diagnosis and reducing related deaths and unnecessary biopsies. This dataset contained 71,066 images with 24 skin disease classes [3], but only 11 of the 24 classes had over 100 images. The images were organized by diagnostic attributes, such as benign, malignant, and disease classes, and clinical attributes, such as patient and skin lesion information, and image type. Due to the diversity and quantity of images contained, this dataset was used to implement the ISIC challenges from 2016–2020 and dramatically contributed to the development of automatic lesion segmentation, lesion attribute detection, and disease classification [3].
The first ISIC 2016 dataset had 900 images for training and 350 images for testing under 2 classes: melanoma and benign. The dataset gradually increased to cover more disease classes from 2017 to 2020. Images in ISIC 2016–2017 were fully paired with the disease annotation from experts, as well as the ground truth of the skin lesion in the form of binary masks. The unique information included in the ISIC dataset is the diameter of each skin lesion, which can help to clarify the stage of melanoma. In the ISIC 2018 challenge, the labeled HAM 10,000 dataset served as the training set, which included 10,015 training images with uneven distribution from 7 classes. The BCN 20,000 dataset was utilized as labeled samples in the ISIC 2019 and ISIC 2020 challenge, which consisted of 19,424 dermoscopic images from 9 classes. It is worth noting that not all ISIC datasets were completely labeled in the ISIC 2018–2020 datasets.
To prompt research into diverse skin diseases, images from more disease categories have been (and continue to be) collected. Dermofit has 1300 macroscopic skin lesion images with corresponding segmented masks over 10 classes of diseases, which were captured by a camera under standardized conditions for quality control [14]. DermNet covers 23 classes of skin disease with 21,844 clinical images [11]. For precise diagnosis, melanoma and nevus can be further refined into several subtypes. For example, the EDRA dataset contains only 1011 dermoscopic images from 20 specific categories, including 8 categories of nevus, 6 categories of melanoma, and 6 other skin diseases. The images under melanoma were further divided into melanoma, melanoma (in situ), melanoma (less than 0.76 mm), melanoma (0.76 to 1.5 mm), melanoma (more than 1.5 mm), and melanoma metastasis.
Table 1 summarizes these datasets according to image number, disease categories, number of labeled images, and binary segmentation mask inclusion. It can be seen that the ISIC Archive is the largest public repository with expert annotation from 25 types of diseases, and PH2 and Med-Node are smaller datasets with a focus on distinguishing between nevus and melanoma.
Table 1. Summary of publicly available skin lesion datasets.
| Dataset | Image Number | Disease Category | Labeled Images | Segmentation Mask |
|---|---|---|---|---|
| PH2 [8] | 200 | 2 | All | No |
| Med-Node [12] | 170 | 2 | All | No |
| ISIC Archive [3] | 71,066 | 25 | All | No |
| ISIC 2016 [15] | 1279 | 2 | All | Yes |
| ISIC 2017 [16] | 2600 | 3 | All | Yes |
| ISIC 2018 [17] | 11,527 | 7 | 10,015 | Yes |
| ISIC 2019 [18] | 33,569 | 8 | 25,331 | No |
| ISIC 2020 [6] | 44,108 | 9 | 33,126 | No |
| HAM 10,000 [9] | 10,015 | 7 | All | No |
| BCN 20,000 [10] | 19,424 | 8 | All | No |
| EDRA [19] | 1011 | 10 | All | No |
| DermNet [11] | 19,500 | 23 | All | No |
| Dermofit [14] | 1300 | 10 | All | No |
Developing a representative dataset for accurate skin diagnosis requires tremendous manpower and time. Hence, it is common to see that one dataset is utilized for multiple diagnosis tasks. For example, the ISIC datasets can be used to identify melanoma or classify diverse skin lesions, while some types of skin lesions have more representative samples than others. Such an uneven data distribution may hinder the ability of developed diagnostic models to handle a desired variety of tasks.
There are no clear guidelines or criteria on the dataset selection for a specific diagnosis task; moreover, there is little discussion on the required size and distribution of a dataset. A small dataset may lead to insufficient model learning, and a large dataset may result in a high computational workload. The trade-off between the size and distribution of the dataset and the accuracy of the diagnosis model should be addressed. On the other hand, it is hard to identify diseases by a diagnosis model for unseen diseases, or diseases with similar symptoms. These intricate factors may restrict the accuracy and, hence, adoption of diagnosis models developed using publicly available datasets in clinical diagnoses.
2.2. Image Preprocessing
Hair, illumination, circles, and black frames can often be found in images in the ISIC datasets, as seen in Figure 2. Their presence lowers image quality and, therefore, hinders subsequent analysis. To alleviate these factors, image pre-processing is utilized, which can involve any combination or all of the following: resizing, grayscale conversion, noise removal, and contrast enhancement. These methods generally improve the diagnosis efficiency and effectiveness, as well as segmentation and classification processes.
Image resizing is a general go-to solution for managing images with varied sizes. With this method, most images can be effectively resized to similar dimensions. Moreover, DL methods can handle images with a broad range of sizes. For example, CNNs have translation invariance, which means that the trained model can accommodate varied image sizes without sacrificing performance [20]. Thus, researchers can confidently tackle skin classification and segmentation tasks under diverse image scales. Resizing images involves cropping irrelevant parts of images and standardizing the pixel count before extracting the region of interest (ROI). This can shorten the computational time and simplify the following diagnosis tasks. The resized images are often converted to grayscale images using grayscale conversion to minimize the influence caused by different skin colors [21]. To eliminate various noise artifacts, as shown in Figure 3, various filters can be introduced [22]. A monomorphic filter can normalize image brightness and increase contrast for ROI determination. Hair removal filters are selected according to hair characteristics. A Gaussian filter, average filter, and median filter can be utilized to remove thin hairs, while an inpainting method can be used for thick hairs and a Dull Razor method for more hair [23][24].
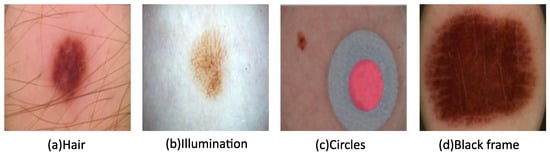
Figure 3. Images with various types of noise from the ISIC archive.
Some ISIC images have colored circles, as shown in Figure 3c, which can be detected and then refilled with healthy skin. The black framing seen in the corners of Figure 3d is often replaced using morphological region filling. Following noise removal, contrast enhancement is achieved for the ROI determination using a bottom-hat filter and contrast-limited adaptive histogram equalization. Face parsing is another commonly used technique to remove the eyes, eyebrows, nose, and mouth as part of the pre-processing process prior to facial skin lesion analysis [25].
2.3. Segmentation and Classification Evaluation Metrics
The performance of skin lesion segmentation and classification can be evaluated by precision, recall (sensitivity), specificity, accuracy, and F1-score, which are from Equation (1) to Equation (5) [26].
where NTP, NTN, NFP, and NFN are defined as the number of true-positive samples under each class, true-negative samples under each class, false-positive samples under each class (pixels with wrongly detected class), and false-negative samples (pixels wrongly detected as other classes), respectively.
Other metrics for classification performance measurements include AUC and IoU. AUC stands for the area under the ROC (receiver operating characteristic) curve, and a higher AUC refers to a better distinguishing capability. Intersection over union (IoU) is a good metric for measuring the overlap between two bounding boxes or masks [27]. Table 2 provides a summary of the segmentation and classification tasks, DL methods, and the corresponding evaluation metrics.
Table 2. Summary of tasks, DL methods, and the evaluation metrics. Jac: Jaccard index, Acc: accuracy, FS: F1-score, SP: specificity, SS: sensitivity.
| Task | DL Methods | Metrics | Ref | |||||
|---|---|---|---|---|---|---|---|---|
| Jac | Acc | FS | SP | SS | Dice | |||
| Skin Lesion Segmentation | SkinNet | √ | √ | √ | [28] | |||
| Skin Lesion Segmentation | FrCN | √ | √ | √ | [29] | |||
| Skin Lesion Segmentation | YOLO and Grabcut Algorithm |
√ | √ | √ | √ | [30] | ||
| Skin Lesion Segmentation and Classification |
Swarm Intelligence (SI) | √ | √ | √ | √ | √ | √ | [31] |
| Skin Lesion Segmentation | UNet and unsupervised approach |
√ | √ | [32] | ||||
| Skin Lesion Segmentation | CNN and Transformer | √ | [20] | |||||
| Skin Lesion Classification | VGG and Inception V3 | √ | √ | [33] | ||||
| Skin Lesion Classification | CNN and Transfer Learning | √ | √ | √ | √ | [34] | ||
| Melanoma Classification | ResNet and SVM | √ | [35] | |||||
| Melanoma Classification | Fast RCNN and DenseNet | √ | [36] | |||||
References
- ALKolifi-ALEnezi, N.S. A Method Of Skin Disease Detection Using Image Processing And Machine Learning. Procedia Comput. Sci. 2019, 163, 85–92.
- Skin Disorders: Pictures, Causes, Symptoms, and Treatment. Available online: https://www.healthline.com/health/skin-disorders (accessed on 21 February 2023).
- ISIC Archive. Available online: https://www.isic-archive.com/#!/topWithHeader/wideContentTop/main (accessed on 20 February 2023).
- Sun, J.; Yao, K.; Huang, K.; Huang, D. Machine learning applications in scaffold based bioprinting. Mater. Today Proc. 2022, 70, 17–23.
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842.
- Rotemberg, V.; Kurtansky, N.; Betz-Stablein, B.; Caffery, L.; Chousakos, E.; Codella, N.; Combalia, M.; Dusza, S.; Guitera, P.; Gutman, D.; et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci. Data 2021, 8, 34.
- Melanoma Skin Cancer Rreport. Melanoma UK. 2020. Available online: https://www.melanomauk.org.uk/2020-melanoma-skin-cancer-report (accessed on 20 February 2023).
- Mendonça, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.; Rozeira, J. PH 2-A dermoscopic image database for research and benchmarking. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440.
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 1–9.
- Combalia, M.; Codella, N.C.; Rotemberg, V.; Helba, B.; Vilaplana, V.; Reiter, O.; Carrera, C.; Barreiro, A.; Halpern, A.C.; Puig, S.; et al. Bcn20000: Dermoscopic lesions in the wild. arXiv 2019, arXiv:1908.02288.
- Dermnet. Kaggle. Available online: https://www.kaggle.com/datasets/shubhamgoel27/dermnet (accessed on 20 February 2023).
- Giotis, I.; Molders, N.; Land, S.; Biehl, M.; Jonkman, M.F.; Petkov, N. MED-NODE: A computer-assisted melanoma diagnosis system using non-dermoscopic images. Expert Syst. Appl. 2015, 42, 6578–6585.
- Yap, J.; Yolland, W.; Tschandl, P. Multimodal skin lesion classification using deep learning. Exp. Dermatol. 2018, 27, 1261–1267.
- Dermofit Image Library Available from The University of Edinburgh. Available online: https://licensing.edinburgh-innovations.ed.ac.uk/product/dermofit-image-library (accessed on 20 February 2023).
- Gutman, D.; Codella, N.C.; Celebi, E.; Helba, B.; Marchetti, M.; Mishra, N.; Halpern, A. Skin lesion analysis toward melanoma detection: A challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC). arXiv 2016, arXiv:1605.01397.
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging, hosted by the international skin imaging collaboration. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172.
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv 2019, arXiv:1902.03368.
- ISIC Challenge. Available online: https://challenge.isic-archive.com/landing/2019/ (accessed on 21 February 2023).
- Kawahara, J.; Daneshvar, S.; Argenziano, G.; Hamarneh, G. Seven-point checklist and skin lesion classification using multitask multimodal neural nets. IEEE J. Biomed. Health Inform. 2019, 23, 538–546.
- Alahmadi, M.D.; Alghamdi, W. Semi-Supervised Skin Lesion Segmentation With Coupling CNN and Transformer Features. IEEE Access 2022, 10, 122560–122569.
- Abhishek, K.; Hamarneh, G.; Drew, M.S. Illumination-based transformations improve skin lesion segmentation in dermoscopic images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 728–729.
- Oliveira, R.B.; Mercedes Filho, E.; Ma, Z.; Papa, J.P.; Pereira, A.S.; Tavares, J.M.R. Computational methods for the image segmentation of pigmented skin lesions: A review. Comput. Methods Programs Biomed. 2016, 131, 127–141.
- Hameed, N.; Shabut, A.M.; Ghosh, M.K.; Hossain, M.A. Multi-class multi-level classification algorithm for skin lesions classification using machine learning techniques. Expert Syst. Appl. 2020, 141, 112961.
- Toossi, M.T.B.; Pourreza, H.R.; Zare, H.; Sigari, M.H.; Layegh, P.; Azimi, A. An effective hair removal algorithm for dermoscopy images. Ski. Res. Technol. 2013, 19, 230–235.
- Zhang, C.; Huang, G.; Yao, K.; Leach, M.; Sun, J.; Huang, K.; Zhou, X.; Yuan, L. A Comparison of Applying Image Processing and Deep Learning in Acne Region Extraction. J. Image Graph. 2022, 10, 1–6.
- Shetty, B.; Fernandes, R.; Rodrigues, A.P.; Chengoden, R.; Bhattacharya, S.; Lakshmanna, K. Skin lesion classification of dermoscopic images using machine learning and convolutional neural network. Sci. Rep. 2022, 12, 18134.
- Gulzar, Y.; Khan, S.A. Skin Lesion Segmentation Based on Vision Transformers and Convolutional Neural Networks—A Comparative Study. Appl. Sci. 2022, 12, 5990.
- Vesal, S.; Ravikumar, N.; Maier, A. SkinNet: A deep learning framework for skin lesion segmentation. In Proceedings of the 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC), Sydney, Australia, 10–17 November 2018; pp. 1–3.
- Al-Masni, M.A.; Al-Antari, M.A.; Choi, M.T.; Han, S.M.; Kim, T.S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 2018, 162, 221–231.
- Ünver, H.M.; Ayan, E. Skin lesion segmentation in dermoscopic images with combination of YOLO and grabcut algorithm. Diagnostics 2019, 9, 72.
- Thapar, P.; Rakhra, M.; Cazzato, G.; Hossain, M.S. A novel hybrid deep learning approach for skin lesion segmentation and classification. J. Healthc. Eng. 2022, 2022, 1709842.
- Ali, A.R.; Li, J.; Trappenberg, T. Supervised versus unsupervised deep learning based methods for skin lesion segmentation in dermoscopy images. In Proceedings of the Advances in Artificial Intelligence: 32nd Canadian Conference on Artificial Intelligence, Canadian AI 2019, Kingston, ON, Canada, 28–31 May 2019; pp. 373–379.
- Gupta, S.; Panwar, A.; Mishra, K. Skin disease classification using dermoscopy images through deep feature learning models and machine learning classifiers. In Proceedings of the IEEE EUROCON 2021–19th International Conference on Smart Technologies, Lviv, Ukraine, 6–8 July 2021; pp. 170–174.
- Kassem, M.A.; Hosny, K.M.; Fouad, M.M. Skin lesions classification into eight classes for ISIC 2019 using deep convolutional neural network and transfer learning. IEEE Access 2020, 8, 114822–114832.
- Khan, M.A.; Javed, M.Y.; Sharif, M.; Saba, T.; Rehman, A. Multi-model deep neural network based features extraction and optimal selection approach for skin lesion classification. In Proceedings of the 2019 International Conference on Computer and Information Sciences (ICCIS), Aljouf, Saudi Arabia, 3–4 April 2019; pp. 1–7.
- Khan, M.A.; Sharif, M.; Akram, T.; Bukhari, S.A.C.; Nayak, R.S. Developed Newton-Raphson based deep features selection framework for skin lesion recognition. Pattern Recognit. Lett. 2020, 129, 293–303.
More
Information
Subjects:
Engineering, Biomedical
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
4.2K
Revisions:
3 times
(View History)
Update Date:
18 Apr 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No