
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Upendar Rao Golla | -- | 3831 | 2023-03-06 13:49:59 | | | |
| 2 | Lindsay Dong | Meta information modification | 3831 | 2023-03-07 07:36:35 | | |
Video Upload Options
The organization of eukaryotic genome in the nucleus, a double-membraned organelle separated from the cytoplasm, is highly complex and dynamic. The functional architecture of the nucleus is confined by the layers of internal and cytoplasmic elements, including chromatin organization, nuclear envelope associated proteome and transport, nuclear–cytoskeletal contacts, and the mechano-regulatory signaling cascades. The size and morphology of the nucleus could impose a significant impact on nuclear mechanics, chromatin organization, gene expression, cell functionality and disease development. The maintenance of nuclear organization during genetic or physical perturbation is crucial for the viability and lifespan of the cell. Abnormal nuclear envelope morphologies, such as invagination and blebbing, have functional implications in several human disorders, including cancer, accelerated aging, thyroid disorders, and different types of neuro-muscular diseases.
1. Introduction
2. Contribution of Nuclear Constituents in Regulation of Nuclear Morphology
2.1. Nuclear Envelope Proteins
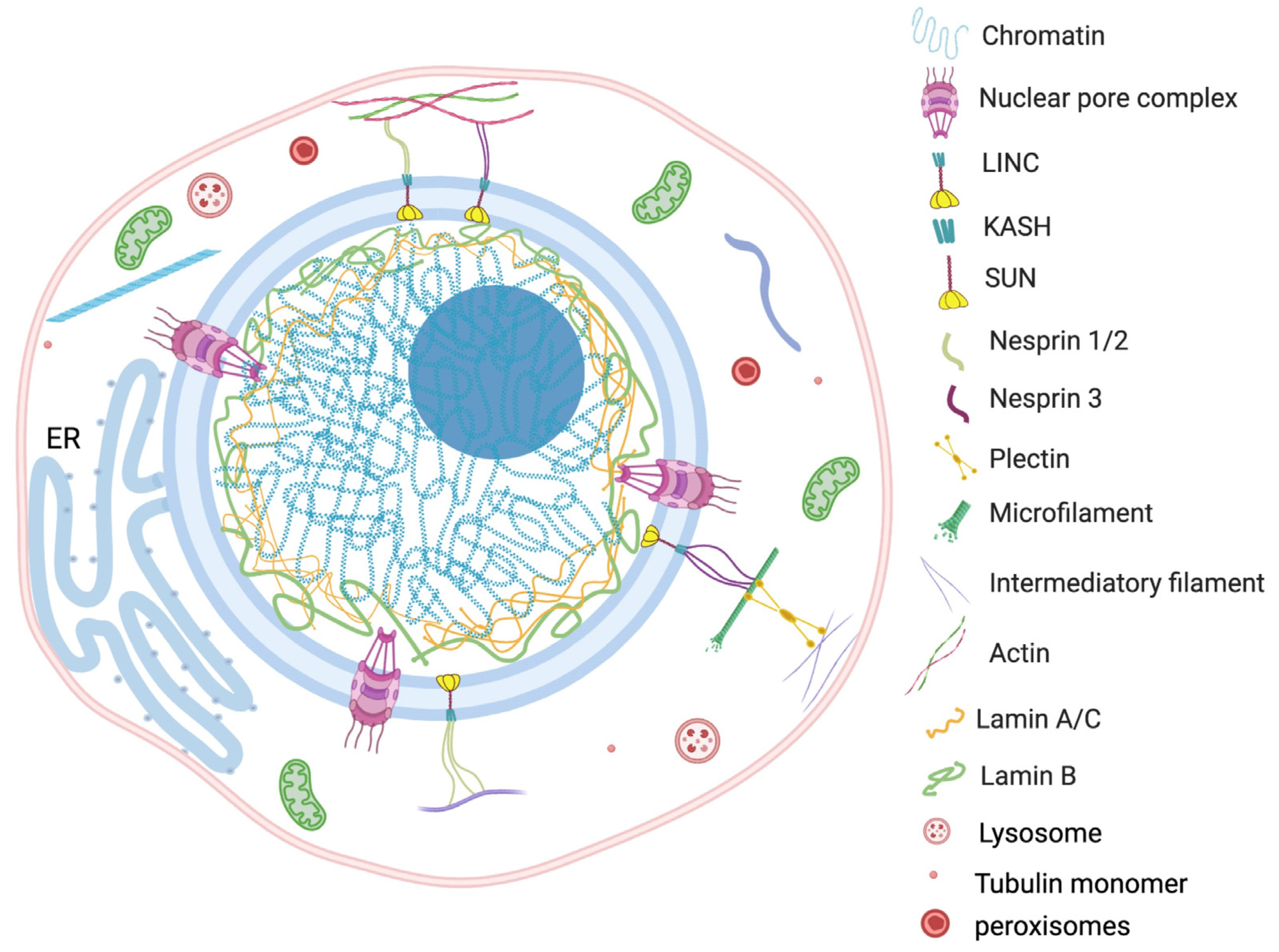
Nuclear envelopes are the structural and physiological interface between the central genomic material and cytoplasm of the cell. The double lipid membrane of the nuclear envelope originates from ER and remains in continuous contact with its network afterward. In contrast to the origin, both the outer nuclear membrane (ONM) and inner nuclear membrane (INM) of the nuclear envelope are enriched with a very distinguished set of proteomes (Figure 1) [14]. These subsets of proteins play key roles in bidirectional nucleoplasmic transportation, maintenance of nuclear architecture, cell cycle control, chromatin organization, gene regulation and DNA repair. The most complex macromolecular assemblies of the nuclear envelope are nuclear pore complexes (NPCs) [15][16][17][18]. NPCs encompass multiple subsets of more than 30 types of nuclear pore proteins called nucleoporins (Nups) [19][20]. The de novo assembly of Nups during interphase starts with the accumulation of Nups in both the outer and inner nuclear membranes, and the subsequent fusion these proteins forms the doughnut-shaped core (consisting of eight spokes arranged around a central channel) of NPC [21][22][23]. The fusion creates an energetically unfavorable and highly curved membrane that surrounds the NPC [24].

2.2. Nuclear Membrane Composition
2.3. Genome Organization
2.4. Nuclear Subcompartments and Nucleolus
2.5. Nucleus and Cytoplasmic Components
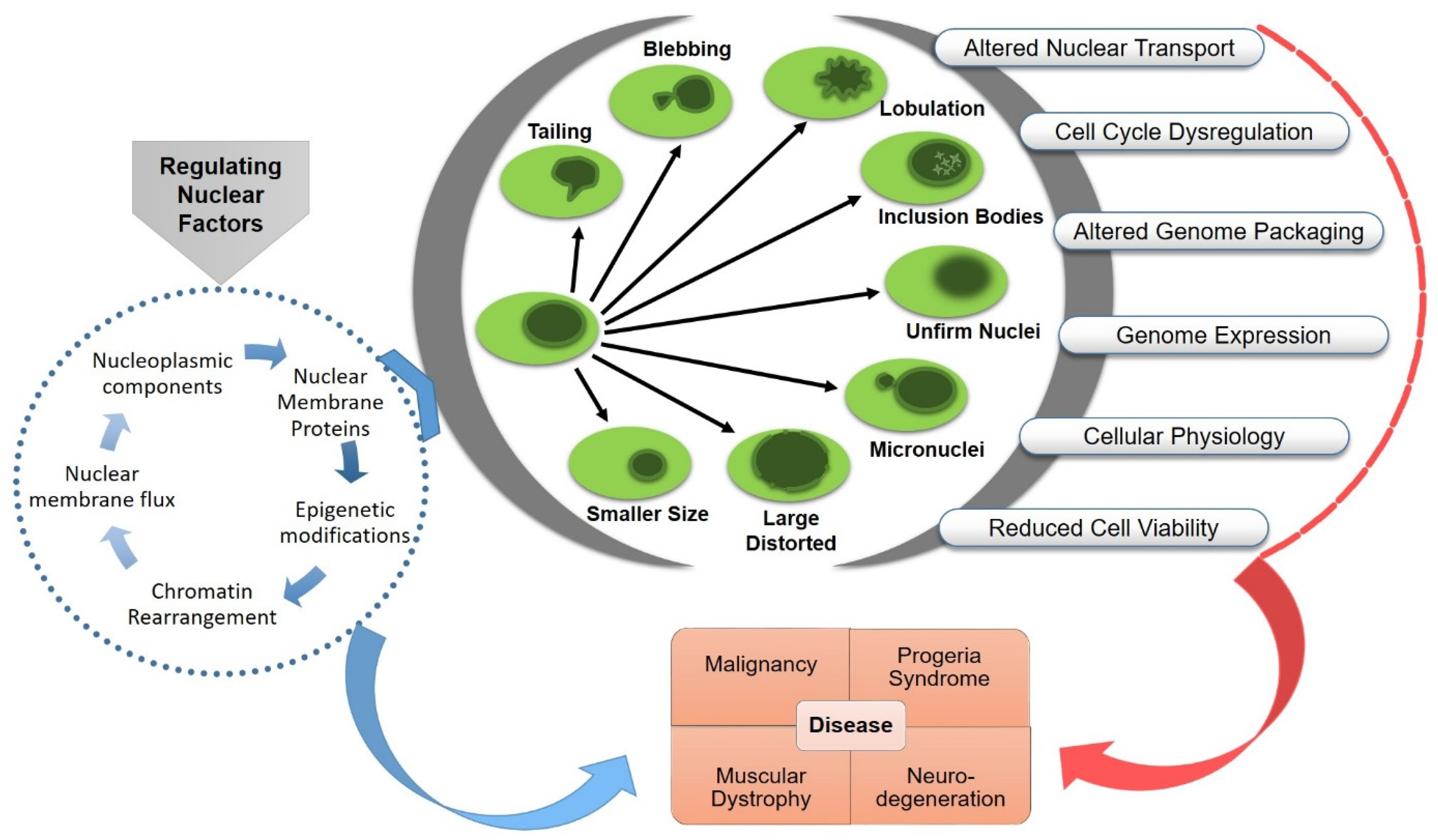
3. Functional Consequences of Nuclear Aberrations
4. Therapeutic Approaches Targeting Nucleus in Disease and Identification of Potential Diagnostic Biomarkers
References
- Mukherjee, R.N.; Chen, P.; Levy, D.L. Recent advances in understanding nuclear size and shape. Nucleus 2016, 7, 167–186.
- Stephens, A.D.; Liu, P.Z.; Banigan, E.J.; Almassalha, L.M.; Backman, V.; Adam, S.A.; Goldman, R.D.; Marko, J.F. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell 2018, 29, 220–233.
- Hobson, C.M.; Kern, M.; O’Brien, E.T., 3rd; Stephens, A.D.; Falvo, M.R.; Superfine, R. Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics. Mol. Biol. Cell 2020, 31, 1788–1801.
- Walters, A.D.; Bommakanti, A.; Cohen-Fix, O. Shaping the nucleus: Factors and forces. J. Cell. Biochem. 2012, 113, 2813–2821.
- Anding, A.L.; Baehrecke, E.H. Cleaning House: Selective Autophagy of Organelles. Dev. Cell 2017, 41, 10–22.
- Hiepen, C.; Mendez, P.L.; Knaus, P. It Takes Two to Tango: Endothelial TGFbeta/BMP Signaling Crosstalk with Mechanobiology. Cells 2020, 9, 1965.
- Lord, C.L.; Wente, S.R. Nuclear envelope-vacuole contacts mitigate nuclear pore complex assembly stress. J. Cell Biol. 2020, 219, e202001165.
- Skinner, B.M.; Johnson, E.E. Nuclear morphologies: Their diversity and functional relevance. Chromosoma 2017, 126, 195–212.
- Ramdas, N.M.; Shivashankar, G.V. Cytoskeletal control of nuclear morphology and chromatin organization. J. Mol. Biol. 2015, 427, 695–706.
- Uhler, C.; Shivashankar, G.V. Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Biol. 2017, 18, 717–727.
- Aureille, J.; Buffiere-Ribot, V.; Harvey, B.E.; Boyault, C.; Pernet, L.; Andersen, T.; Bacola, G.; Balland, M.; Fraboulet, S.; Van Landeghem, L.; et al. Nuclear envelope deformation controls cell cycle progression in response to mechanical force. EMBO Rep. 2019, 20, e48084.
- Stowers, R.S.; Shcherbina, A.; Israeli, J.; Gruber, J.J.; Chang, J.; Nam, S.; Rabiee, A.; Teruel, M.N.; Snyder, M.P.; Kundaje, A.; et al. Matrix stiffness induces a tumorigenic phenotype in mammary epithelium through changes in chromatin accessibility. Nat. Biomed. Eng. 2019, 3, 1009–1019.
- Kussick, S.J.; Stirewalt, D.L.; Yi, H.S.; Sheets, K.M.; Pogosova-Agadjanyan, E.; Braswell, S.; Norwood, T.H.; Radich, J.P.; Wood, B.L. A distinctive nuclear morphology in acute myeloid leukemia is strongly associated with loss of HLA-DR expression and FLT3 internal tandem duplication. Leukemia 2004, 18, 1591–1598.
- Garapati, H.S.; Mishra, K. Comparative genomics of nuclear envelope proteins. BMC Genom. 2018, 19, 823.
- Alber, F.; Dokudovskaya, S.; Veenhoff, L.M.; Zhang, W.; Kipper, J.; Devos, D.; Suprapto, A.; Karni-Schmidt, O.; Williams, R.; Chait, B.T.; et al. Determining the architectures of macromolecular assemblies. Nature 2007, 450, 683–694.
- Alber, F.; Dokudovskaya, S.; Veenhoff, L.M.; Zhang, W.; Kipper, J.; Devos, D.; Suprapto, A.; Karni-Schmidt, O.; Williams, R.; Chait, B.T.; et al. The molecular architecture of the nuclear pore complex. Nature 2007, 450, 695–701.
- Devos, D.; Dokudovskaya, S.; Alber, F.; Williams, R.; Chait, B.T.; Sali, A.; Rout, M.P. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004, 2, e380.
- Dultz, E.; Wojtynek, M.; Medalia, O.; Onischenko, E. The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth. Cells 2022, 11, 1456.
- Fernandez-Martinez, J.; Kim, S.J.; Shi, Y.; Upla, P.; Pellarin, R.; Gagnon, M.; Chemmama, I.E.; Wang, J.; Nudelman, I.; Zhang, W.; et al. Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell 2016, 167, 1215–1228 e1225.
- Kim, S.J.; Fernandez-Martinez, J.; Nudelman, I.; Shi, Y.; Zhang, W.; Raveh, B.; Herricks, T.; Slaughter, B.D.; Hogan, J.A.; Upla, P.; et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature 2018, 555, 475–482.
- Fernandez-Martinez, J.; Rout, M.P. Nuclear pore complex biogenesis. Curr. Opin Cell Biol. 2009, 21, 603–612.
- Devos, D.; Dokudovskaya, S.; Williams, R.; Alber, F.; Eswar, N.; Chait, B.T.; Rout, M.P.; Sali, A. Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2006, 103, 2172–2177.
- Otsuka, S.; Ellenberg, J. Mechanisms of nuclear pore complex assembly—Two different ways of building one molecular machine. FEBS Lett. 2018, 592, 475–488.
- Hetzer, M.W. The nuclear envelope. Cold Spring Harb Perspect. Biol. 2010, 2, a000539.
- Thorpe, S.D.; Lee, D.A. Dynamic regulation of nuclear architecture and mechanics-a rheostatic role for the nucleus in tailoring cellular mechanosensitivity. Nucleus 2017, 8, 287–300.
- Hao, H.; Starr, D.A. SUN/KASH interactions facilitate force transmission across the nuclear envelope. Nucleus 2019, 10, 73–80.
- Hieda, M. Signal Transduction across the Nuclear Envelope: Role of the LINC Complex in Bidirectional Signaling. Cells 2019, 8, 124.
- Campbell, J.L.; Lorenz, A.; Witkin, K.L.; Hays, T.; Loidl, J.; Cohen-Fix, O. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol. Biol. Cell 2006, 17, 1768–1778.
- Barbosa, A.D.; Sembongi, H.; Su, W.M.; Abreu, S.; Reggiori, F.; Carman, G.M.; Siniossoglou, S. Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol. Biol. Cell 2015, 26, 3641–3657.
- Friederichs, J.M.; Ghosh, S.; Smoyer, C.J.; McCroskey, S.; Miller, B.D.; Weaver, K.J.; Delventhal, K.M.; Unruh, J.; Slaughter, B.D.; Jaspersen, S.L. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet. 2011, 7, e1002365.
- Sosa Ponce, M.L.; Moradi-Fard, S.; Zaremberg, V.; Cobb, J.A. SUNny Ways: The Role of the SUN-Domain Protein Mps3 Bridging Yeast Nuclear Organization and Lipid Homeostasis. Front. Genet. 2020, 11, 136.
- Loewen, C.J.; Gaspar, M.L.; Jesch, S.A.; Delon, C.; Ktistakis, N.T.; Henry, S.A.; Levine, T.P. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 2004, 304, 1644–1647.
- Briand, N.; Collas, P. Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol. 2020, 21, 85.
- Nava, M.M.; Miroshnikova, Y.A.; Biggs, L.C.; Whitefield, D.B.; Metge, F.; Boucas, J.; Vihinen, H.; Jokitalo, E.; Li, X.; Garcia Arcos, J.M.; et al. Heterochromatin-Driven Nuclear Softening Protects the Genome against Mechanical Stress-Induced Damage. Cell 2020, 181, 800–817.e822.
- Imbalzano, K.M.; Cohet, N.; Wu, Q.; Underwood, J.M.; Imbalzano, A.N.; Nickerson, J.A. Nuclear shape changes are induced by knockdown of the SWI/SNF ATPase BRG1 and are independent of cytoskeletal connections. PLoS ONE 2013, 8, e55628.
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306.
- Morelli, F.F.; Verbeek, D.S.; Bertacchini, J.; Vinet, J.; Mediani, L.; Marmiroli, S.; Cenacchi, G.; Nasi, M.; De Biasi, S.; Brunsting, J.F.; et al. Aberrant Compartment Formation by HSPB2 Mislocalizes Lamin A and Compromises Nuclear Integrity and Function. Cell Rep. 2017, 20, 2100–2115.
- Farley, K.I.; Surovtseva, Y.; Merkel, J.; Baserga, S.J. Determinants of mammalian nucleolar architecture. Chromosoma 2015, 124, 323–331.
- Matos-Perdomo, E.; Machin, F. Nucleolar and Ribosomal DNA Structure under Stress: Yeast Lessons for Aging and Cancer. Cells 2019, 8, 779.
- Penzo, M.; Montanaro, L.; Trere, D.; Derenzini, M. The Ribosome Biogenesis-Cancer Connection. Cells 2019, 8, 55.
- Witkin, K.L.; Chong, Y.; Shao, S.; Webster, M.T.; Lahiri, S.; Walters, A.D.; Lee, B.; Koh, J.L.; Prinz, W.A.; Andrews, B.J.; et al. The budding yeast nuclear envelope adjacent to the nucleolus serves as a membrane sink during mitotic delay. Curr. Biol. 2012, 22, 1128–1133.
- Matsumoto, A.; Sakamoto, C.; Matsumori, H.; Katahira, J.; Yasuda, Y.; Yoshidome, K.; Tsujimoto, M.; Goldberg, I.G.; Matsuura, N.; Nakao, M.; et al. Loss of the integral nuclear envelope protein SUN1 induces alteration of nucleoli. Nucleus 2016, 7, 68–83.
- Sen Gupta, A.; Sengupta, K. Lamin B2 Modulates Nucleolar Morphology, Dynamics, and Function. Mol. Cell. Biol. 2017, 37, e00274-17.
- Bravo-Sagua, R.; Torrealba, N.; Paredes, F.; Morales, P.E.; Pennanen, C.; Lopez-Crisosto, C.; Troncoso, R.; Criollo, A.; Chiong, M.; Hill, J.A.; et al. Organelle communication: Signaling crossroads between homeostasis and disease. Int. J. Biochem. Cell Biol. 2014, 50, 55–59.
- Dakik, P.; Titorenko, V.I. Communications between Mitochondria, the Nucleus, Vacuoles, Peroxisomes, the Endoplasmic Reticulum, the Plasma Membrane, Lipid Droplets, and the Cytosol during Yeast Chronological Aging. Front. Genet. 2016, 7, 177.
- Kakimoto, Y.; Tashiro, S.; Kojima, R.; Morozumi, Y.; Endo, T.; Tamura, Y. Visualizing multiple inter-organelle contact sites using the organelle-targeted split-GFP system. Sci. Rep. 2018, 8, 6175.
- Monroy-Ramirez, H.C.; Basurto-Islas, G.; Mena, R.; Cisneros, B.; Binder, L.I.; Avila, J.; Garcia-Sierra, F. Alterations in the nuclear architecture produced by the overexpression of tau protein in neuroblastoma cells. J. Alzheimer’s Dis. 2013, 36, 503–520.
- Colon-Bolea, P.; Garcia-Gomez, R.; Shackleton, S.; Crespo, P.; Bustelo, X.R.; Casar, B. RAC1 induces nuclear alterations through the LINC complex to enhance melanoma invasiveness. Mol. Biol. Cell 2020, 31, 2768–2778.
- Lu, W.; Schneider, M.; Neumann, S.; Jaeger, V.M.; Taranum, S.; Munck, M.; Cartwright, S.; Richardson, C.; Carthew, J.; Noh, K.; et al. Nesprin interchain associations control nuclear size. Cell. Mol. Life Sci. 2012, 69, 3493–3509.
- Denais, C.; Lammerding, J. Nuclear mechanics in cancer. Adv. Exp. Med. Biol. 2014, 773, 435–470.
- Meinke, P.; Schirmer, E.C. LINC’ing form and function at the nuclear envelope. FEBS Lett. 2015, 589, 2514–2521.
- Frost, B.; Bardai, F.H.; Feany, M.B. Lamin Dysfunction Mediates Neurodegeneration in Tauopathies. Curr. Biol. 2016, 26, 129–136.
- Essawy, N.; Samson, C.; Petitalot, A.; Moog, S.; Bigot, A.; Herrada, I.; Marcelot, A.; Arteni, A.A.; Coirault, C.; Zinn-Justin, S. An Emerin LEM-Domain Mutation Impairs Cell Response to Mechanical Stress. Cells 2019, 8, 570.
- Dey, P. Cancer nucleus: Morphology and beyond. Diagn. Cytopathol. 2010, 38, 382–390.
- Paramanandam, M.; O’Byrne, M.; Ghosh, B.; Mammen, J.J.; Manipadam, M.T.; Thamburaj, R.; Pakrashi, V. Automated Segmentation of Nuclei in Breast Cancer Histopathology Images. PLoS ONE 2016, 11, e0162053.
- Alvarado-Kristensson, M.; Rossello, C.A. The Biology of the Nuclear Envelope and Its Implications in Cancer Biology. Int. J. Mol. Sci. 2019, 20, 2586.
- Capo-chichi, C.D.; Cai, K.Q.; Smedberg, J.; Ganjei-Azar, P.; Godwin, A.K.; Xu, X.X. Loss of A-type lamin expression compromises nuclear envelope integrity in breast cancer. Chin. J. Cancer 2011, 30, 415–425.
- De Vos, W.H.; Houben, F.; Kamps, M.; Malhas, A.; Verheyen, F.; Cox, J.; Manders, E.M.; Verstraeten, V.L.; van Steensel, M.A.; Marcelis, C.L.; et al. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet. 2011, 20, 4175–4186.
- Earle, A.J.; Kirby, T.J.; Fedorchak, G.R.; Isermann, P.; Patel, J.; Iruvanti, S.; Moore, S.A.; Bonne, G.; Wallrath, L.L.; Lammerding, J. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat. Mater. 2020, 19, 464–473.
- Chow, K.H.; Factor, R.E.; Ullman, K.S. The nuclear envelope environment and its cancer connections. Nat. Rev. Cancer 2012, 12, 196–209.
- Fahrenkrog, B.; Martinelli, V.; Nilles, N.; Fruhmann, G.; Chatel, G.; Juge, S.; Sauder, U.; Di Giacomo, D.; Mecucci, C.; Schwaller, J. Expression of Leukemia-Associated Nup98 Fusion Proteins Generates an Aberrant Nuclear Envelope Phenotype. PLoS ONE 2016, 11, e0152321.
- García-Aguirre, I.; Monterrubio-Ledezma, F.; Alamillo-Iniesta, A.; Castro-Obregón, S.; Cisneros, B. Molecular Basis of Progeroid Diseases. In Clinical Genetics and Genomics of Aging; Gomez-Verjan, J.C., Rivero-Segura, N.A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 187–211.
- Bouska, M.; Huang, K.; Kang, P.; Bai, H. Organelle aging: Lessons from model organisms. J. Genet. Genom. 2019, 46, 171–185.
- Cenni, V.; Capanni, C.; Mattioli, E.; Schena, E.; Squarzoni, S.; Bacalini, M.G.; Garagnani, P.; Salvioli, S.; Franceschi, C.; Lattanzi, G. Lamin A involvement in ageing processes. Ageing Res. Rev. 2020, 62, 101073.
- Martins, F.; Sousa, J.; Pereira, C.D.; da Cruz, E.S.O.A.B.; Rebelo, S. Nuclear envelope dysfunction and its contribution to the aging process. Aging Cell 2020, 19, e13143.
- Lee, S.J.; Jung, Y.S.; Yoon, M.H.; Kang, S.M.; Oh, A.Y.; Lee, J.H.; Jun, S.Y.; Woo, T.G.; Chun, H.Y.; Kim, S.K.; et al. Interruption of progerin-lamin A/C binding ameliorates Hutchinson-Gilford progeria syndrome phenotype. J. Clin. Investig. 2016, 126, 3879–3893.
- Goldman, R.D.; Shumaker, D.K.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Gordon, L.B.; Gruenbaum, Y.; Khuon, S.; Mendez, M.; Varga, R.; et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 8963–8968.
- Spear, E.D.; Hsu, E.T.; Nie, L.; Carpenter, E.P.; Hrycyna, C.A.; Michaelis, S. ZMPSTE24 missense mutations that cause progeroid diseases decrease prelamin A cleavage activity and/or protein stability. Dis. Model. Mech. 2018, 11, dmm033670.
- Holzer, K.; Ori, A.; Cooke, A.; Dauch, D.; Drucker, E.; Riemenschneider, P.; Andres-Pons, A.; DiGuilio, A.L.; Mackmull, M.T.; Bassler, J.; et al. Nucleoporin Nup155 is part of the p53 network in liver cancer. Nat. Commun. 2019, 10, 2147.
- Kalinin, A.A.; Allyn-Feuer, A.; Ade, A.; Fon, G.V.; Meixner, W.; Dilworth, D.; Husain, S.S.; de Wet, J.R.; Higgins, G.A.; Zheng, G.; et al. 3D Shape Modeling for Cell Nuclear Morphological Analysis and Classification. Sci. Rep. 2018, 8, 13658.
- Fischer, A.H.; Bardarov, S., Jr.; Jiang, Z. Molecular aspects of diagnostic nucleolar and nuclear envelope changes in prostate cancer. J. Cell. Biochem. 2004, 91, 170–184.
- Carotenuto, P.; Pecoraro, A.; Palma, G.; Russo, G.; Russo, A. Therapeutic Approaches Targeting Nucleolus in Cancer. Cells 2019, 8, 1090.




