
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ilyas Chohra | -- | 1789 | 2023-02-14 12:13:37 | | | |
| 2 | Conner Chen | + 10 word(s) | 1799 | 2023-02-16 03:41:02 | | | | |
| 3 | Conner Chen | + 2 word(s) | 1801 | 2023-02-17 02:04:26 | | |
Video Upload Options
The CHD (chromodomain-helicase-DNA binding) family consists of nine members (CHD1–CHD9). Their structure is characterised by two consecutive chromodomains in the N-terminal region and an SNF2-like (sucrose-non-fermenting (SNF)) ATP-dependent helicase domain positioned in the central region. They recognise and bind nucleosomes to contribute, in many cases, to the formation of heterochromatin typically marked by the presence of methylated histones and other repressive chromatin markers. CHD proteins can bind to methylated histones through their chromodomains and use their helicase activity to remodel the chromatin and contribute to the formation of heterochromatin, a fundamental feature of chromosomes that ensures genomic stability.
1. CHD Family
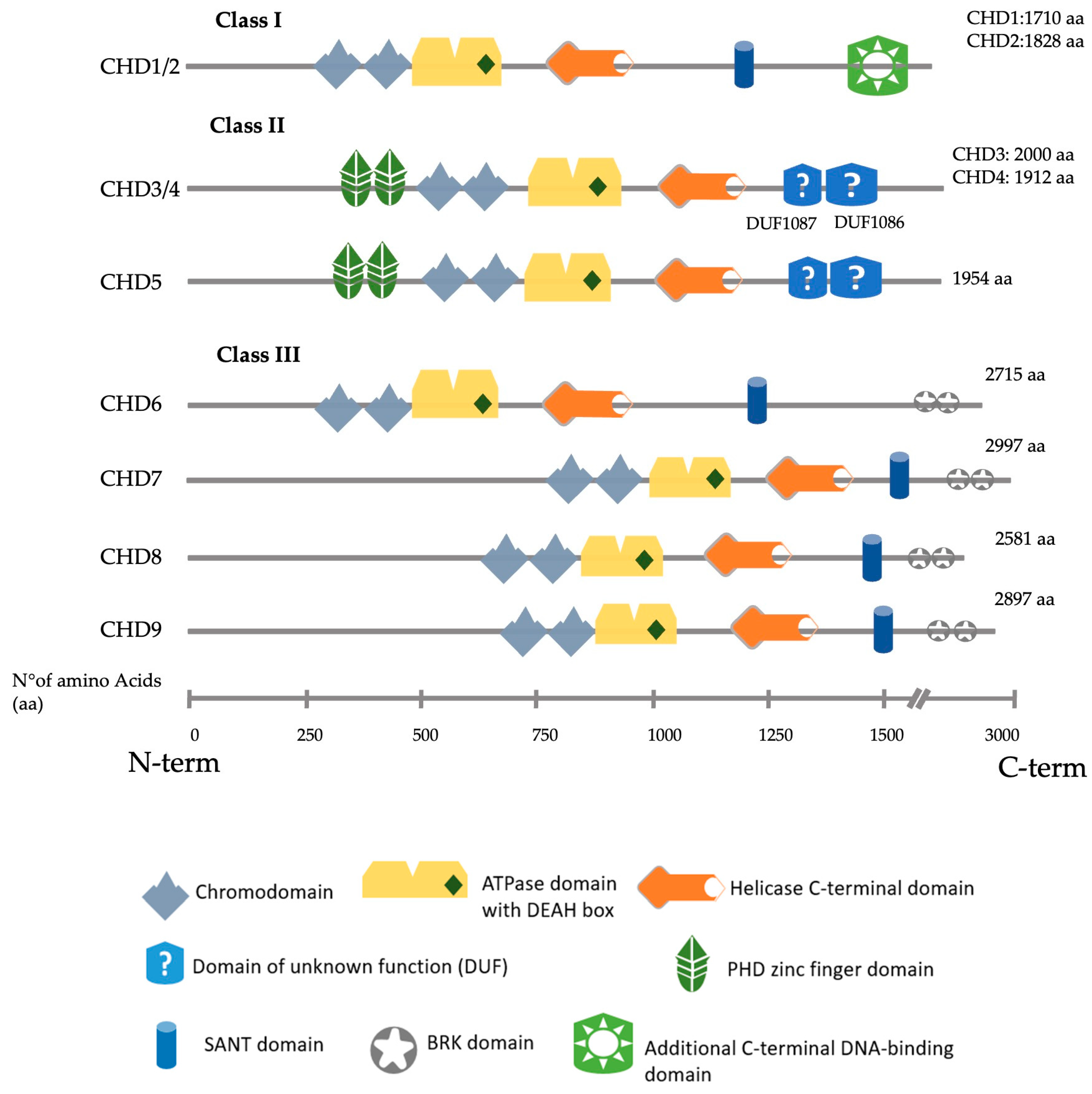
2. CHD3/4
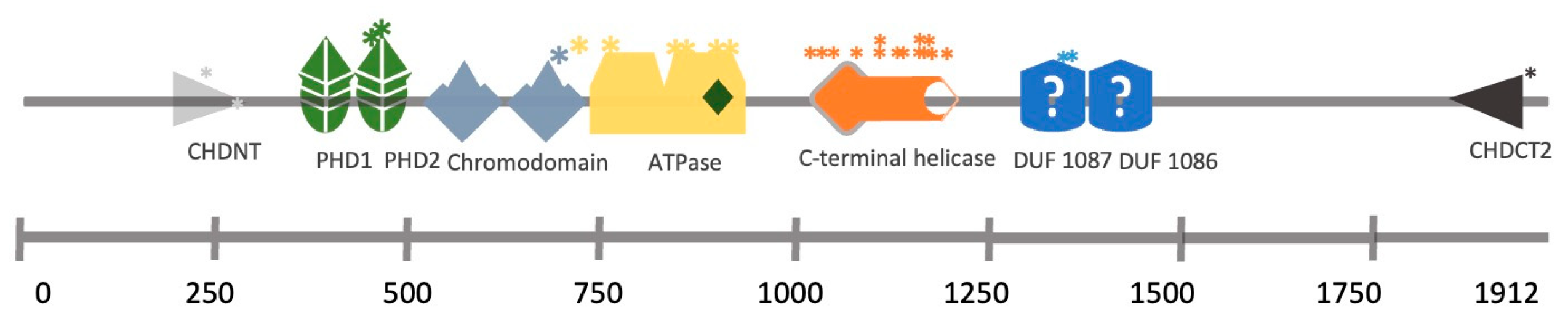
3. CHD7
4. CHD8
References
- Woodage, T.; Basrai, M.A.; Baxevanis, A.D.; Hieter, P.; Collins, F.S. Characterization of the CHD family of proteins. Proc. Natl. Acad. Sci. USA 1997, 94, 11472–11477.
- Marfella, C.G.; Imbalzano, A.N. The Chd family of chromatin remodelers. Mutat. Res. 2007, 618, 30–40.
- Liu, X.R.; Ye, T.T.; Zhang, W.J.; Guo, X.; Wang, J.; Huang, S.P.; Xie, L.S.; Song, X.W.; Deng, W.W.; Li, B.M.; et al. CHD4 variants are associated with childhood idiopathic epilepsy with sinus arrhythmia. CNS Neurosci. Ther. 2021, 27, 1146–1156.
- Allen, M.D.; Religa, T.L.; Freund, S.M.; Bycroft, M. Solution structure of the BRK domains from CHD7. J. Mol. Biol. 2007, 371, 1135–1140.
- Alendar, A.; Berns, A. Sentinels of chromatin: Chromodomain helicase DNA-binding proteins in development and disease. Genes Dev. 2021, 35, 1403–1430.
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489.
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496.
- Blum, M.; Chang, H.Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354.
- Hoffmeister, H.; Fuchs, A.; Erdel, F.; Pinz, S.; Grobner-Ferreira, R.; Bruckmann, A.; Deutzmann, R.; Schwartz, U.; Maldonado, R.; Huber, C.; et al. CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res. 2017, 45, 10534–10554.
- Kloet, S.L.; Baymaz, H.I.; Makowski, M.; Groenewold, V.; Jansen, P.W.; Berendsen, M.; Niazi, H.; Kops, G.J.; Vermeulen, M. Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics. FEBS J. 2015, 282, 1774–1785.
- Lai, A.Y.; Wade, P.A. Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer 2011, 11, 588–596.
- Tong, J.K.; Hassig, C.A.; Schnitzler, G.R.; Kingston, R.E.; Schreiber, S.L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 1998, 395, 917–921.
- Zhao, H.; Han, Z.; Liu, X.; Gu, J.; Tang, F.; Wei, G.; Jin, Y. The chromatin remodeler Chd4 maintains embryonic stem cell identity by controlling pluripotency- and differentiation-associated genes. J. Biol. Chem. 2017, 292, 8507–8519.
- Hirota, A.; Nakajima-Koyama, M.; Ashida, Y.; Nishida, E. The nucleosome remodeling and deacetylase complex protein CHD4 regulates neural differentiation of mouse embryonic stem cells by down-regulating p53. J. Biol. Chem. 2019, 294, 195–209.
- Snijders Blok, L.; Rousseau, J.; Twist, J.; Ehresmann, S.; Takaku, M.; Venselaar, H.; Rodan, L.H.; Nowak, C.B.; Douglas, J.; Swoboda, K.J.; et al. CHD3 helicase domain mutations cause a neurodevelopmental syndrome with macrocephaly and impaired speech and language. Nat. Commun. 2018, 9, 4619.
- Drivas, T.G.; Li, D.; Nair, D.; Alaimo, J.T.; Alders, M.; Altmuller, J.; Barakat, T.S.; Bebin, E.M.; Bertsch, N.L.; Blackburn, P.R.; et al. A second cohort of CHD3 patients expands the molecular mechanisms known to cause Snijders Blok-Campeau syndrome. Eur. J. Hum. Genet. 2020, 28, 1422–1431.
- Weiss, K.; Lazar, H.P.; Kurolap, A.; Martinez, A.F.; Paperna, T.; Cohen, L.; Smeland, M.F.; Whalen, S.; Heide, S.; Keren, B.; et al. The CHD4-related syndrome: A comprehensive investigation of the clinical spectrum, genotype-phenotype correlations, and molecular basis. Genet. Med. 2020, 22, 389–397.
- Farnung, L.; Ochmann, M.; Cramer, P. Nucleosome-CHD4 chromatin remodeler structure maps human disease mutations. Elife 2020, 9, e56178.
- Layman, W.S.; Sauceda, M.A.; Zuo, J. Epigenetic alterations by NuRD and PRC2 in the neonatal mouse cochlea. Hear. Res. 2013, 304, 167–178.
- Nitarska, J.; Smith, J.G.; Sherlock, W.T.; Hillege, M.M.; Nott, A.; Barshop, W.D.; Vashisht, A.A.; Wohlschlegel, J.A.; Mitter, R.; Riccio, A. A Functional Switch of NuRD Chromatin Remodeling Complex Subunits Regulates Mouse Cortical Development. Cell Rep. 2016, 17, 1683–1698.
- Choo, D.I.; Tawfik, K.O.; Martin, D.M.; Raphael, Y. Inner ear manifestations in CHARGE: Abnormalities, treatments, animal models, and progress toward treatments in auditory and vestibular structures. Am. J. Med. Genet. C Semin Med. Genet. 2017, 175, 439–449.
- Van Ravenswaaij-Arts, C.; Martin, D.M. New insights and advances in CHARGE syndrome: Diagnosis, etiologies, treatments, and research discoveries. Am. J. Med. Genet. C Semin Med. Genet. 2017, 175, 397–406.
- Basson, M.A.; van Ravenswaaij-Arts, C. Functional Insights into Chromatin Remodelling from Studies on CHARGE Syndrome. Trends Genet. 2015, 31, 600–611.
- Balendran, V.; Skidmore, J.M.; Ritter, K.E.; Gao, J.; Cimerman, J.; Beyer, L.A.; Hurd, E.A.; Raphael, Y.; Martin, D.M. Chromatin remodeler CHD7 is critical for cochlear morphogenesis and neurosensory patterning. Dev. Biol. 2021, 477, 11–21.
- Bosman, E.A.; Penn, A.C.; Ambrose, J.C.; Kettleborough, R.; Stemple, D.L.; Steel, K.P. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet. 2005, 14, 3463–3476.
- Hurd, E.A.; Capers, P.L.; Blauwkamp, M.N.; Adams, M.E.; Raphael, Y.; Poucher, H.K.; Martin, D.M. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm. Genome 2007, 18, 94–104.
- Ritter, K.E.; Lynch, S.M.; Gorris, A.M.; Beyer, L.A.; Kabara, L.; Dolan, D.F.; Raphael, Y.; Martin, D.M. Loss of the chromatin remodeler CHD7 impacts glial cells and myelination in the mouse cochlear spiral ganglion. Hear. Res. 2022, 426, 108633.
- Ogier, J.M.; Arhatari, B.D.; Carpinelli, M.R.; McColl, B.K.; Wilson, M.A.; Burt, R.A. An intronic mutation in Chd7 creates a cryptic splice site, causing aberrant splicing in a mouse model of CHARGE syndrome. Sci. Rep. 2018, 8, 5482.
- Ogier, J.M.; Carpinelli, M.R.; Arhatari, B.D.; Symons, R.C.; Kile, B.T.; Burt, R.A. CHD7 deficiency in “Looper”, a new mouse model of CHARGE syndrome, results in ossicle malformation, otosclerosis and hearing impairment. PLoS ONE 2014, 9, e97559.
- Hurd, E.A.; Poucher, H.K.; Cheng, K.; Raphael, Y.; Martin, D.M. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development 2010, 137, 3139–3150.
- Ahmed, M.; Moon, R.; Prajapati, R.S.; James, E.; Basson, M.A.; Streit, A. The chromatin remodelling factor Chd7 protects auditory neurons and sensory hair cells from stress-induced degeneration. Commun. Biol. 2021, 4, 1260.
- Micucci, J.A.; Layman, W.S.; Hurd, E.A.; Sperry, E.D.; Frank, S.F.; Durham, M.A.; Swiderski, D.L.; Skidmore, J.M.; Scacheri, P.C.; Raphael, Y.; et al. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum. Mol. Genet. 2014, 23, 434–448.
- Yao, H.; Hill, S.F.; Skidmore, J.M.; Sperry, E.D.; Swiderski, D.L.; Sanchez, G.J.; Bartels, C.F.; Raphael, Y.; Scacheri, P.C.; Iwase, S.; et al. CHD7 represses the retinoic acid synthesis enzyme ALDH1A3 during inner ear development. JCI Insight 2018, 3, e97440.
- Durruthy-Durruthy, R.; Sperry, E.D.; Bowen, M.E.; Attardi, L.D.; Heller, S.; Martin, D.M. Single Cell Transcriptomics Reveal Abnormalities in Neurosensory Patterning of the Chd7 Mutant Mouse Ear. Front. Genet. 2018, 9, 473.
- Koehler, K.R.; Nie, J.; Longworth-Mills, E.; Liu, X.P.; Lee, J.; Holt, J.R.; Hashino, E. Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol. 2017, 35, 583–589.
- Nie, J.; Ueda, Y.; Solivais, A.J.; Hashino, E. CHD7 regulates otic lineage specification and hair cell differentiation in human inner ear organoids. Nat. Commun. 2022, 13, 7053.
- Sugathan, A.; Biagioli, M.; Golzio, C.; Erdin, S.; Blumenthal, I.; Manavalan, P.; Ragavendran, A.; Brand, H.; Lucente, D.; Miles, J.; et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. USA 2014, 111, E4468–E4477.
- Nishiyama, M.; Oshikawa, K.; Tsukada, Y.; Nakagawa, T.; Iemura, S.; Natsume, T.; Fan, Y.; Kikuchi, A.; Skoultchi, A.I.; Nakayama, K.I. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat. Cell Biol. 2009, 11, 172–182.
- Batsukh, T.; Schulz, Y.; Wolf, S.; Rabe, T.I.; Oellerich, T.; Urlaub, H.; Schaefer, I.M.; Pauli, S. Identification and characterization of FAM124B as a novel component of a CHD7 and CHD8 containing complex. PLoS ONE 2012, 7, e52640.
- Batsukh, T.; Pieper, L.; Koszucka, A.M.; von Velsen, N.; Hoyer-Fender, S.; Elbracht, M.; Bergman, J.E.; Hoefsloot, L.H.; Pauli, S. CHD8 interacts with CHD7, a protein which is mutated in CHARGE syndrome. Hum. Mol. Genet. 2010, 19, 2858–2866.
- Hickox, A.E.; Wong, A.C.; Pak, K.; Strojny, C.; Ramirez, M.; Yates, J.R., III; Ryan, A.F.; Savas, J.N. Global Analysis of Protein Expression of Inner Ear Hair Cells. J. Neurosci. 2017, 37, 1320–1339.
- Ding, S.; Lan, X.; Meng, Y.; Yan, C.; Li, M.; Li, X.; Chen, J.; Jiang, W. CHD8 safeguards early neuroectoderm differentiation in human ESCs and protects from apoptosis during neurogenesis. Cell Death Dis. 2021, 12, 981.
- Thompson, B.A.; Tremblay, V.; Lin, G.; Bochar, D.A. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol. Cell Biol. 2008, 28, 3894–3904.
- Nishiyama, M.; Skoultchi, A.I.; Nakayama, K.I. Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-beta-catenin signaling pathway. Mol. Cell Biol. 2012, 32, 501–512.
- DeJonge, R.E.; Liu, X.P.; Deig, C.R.; Heller, S.; Koehler, K.R.; Hashino, E. Modulation of Wnt Signaling Enhances Inner Ear Organoid Development in 3D Culture. PLoS ONE 2016, 11, e0162508.
- Wright, K.D.; Mahoney Rogers, A.A.; Zhang, J.; Shim, K. Cooperative and independent functions of FGF and Wnt signaling during early inner ear development. BMC Dev. Biol. 2015, 15, 33.
- Kawamura, A.; Katayama, Y.; Kakegawa, W.; Ino, D.; Nishiyama, M.; Yuzaki, M.; Nakayama, K.I. The autism-associated protein CHD8 is required for cerebellar development and motor function. Cell Rep. 2021, 35, 108932.





