
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Victor Hugo Antonio Joaquim | -- | 1955 | 2023-02-01 13:23:53 | | | |
| 2 | Jessie Wu | Meta information modification | 1955 | 2023-02-02 02:34:07 | | | | |
| 3 | Jessie Wu | + 2 word(s) | 1957 | 2023-02-02 02:35:42 | | | | |
| 4 | Jessie Wu | + 2 word(s) | 1959 | 2023-02-02 02:44:33 | | |
Video Upload Options
Circular RNAs (circRNAs) are a family of noncoding RNAs (ncRNAs) that are endogenous and widely distributed in different species, performing several functions, mainly their association with microRNAs (miRNAs) and RNA-binding proteins. Cardiovascular diseases (CVDs) remain the leading cause of death worldwide; therefore, the development of new therapies and strategies, such as gene therapies or nonpharmacological therapies, with low cost, such as physical exercise, to alleviate these diseases is of extreme importance for society.
1. Introduction
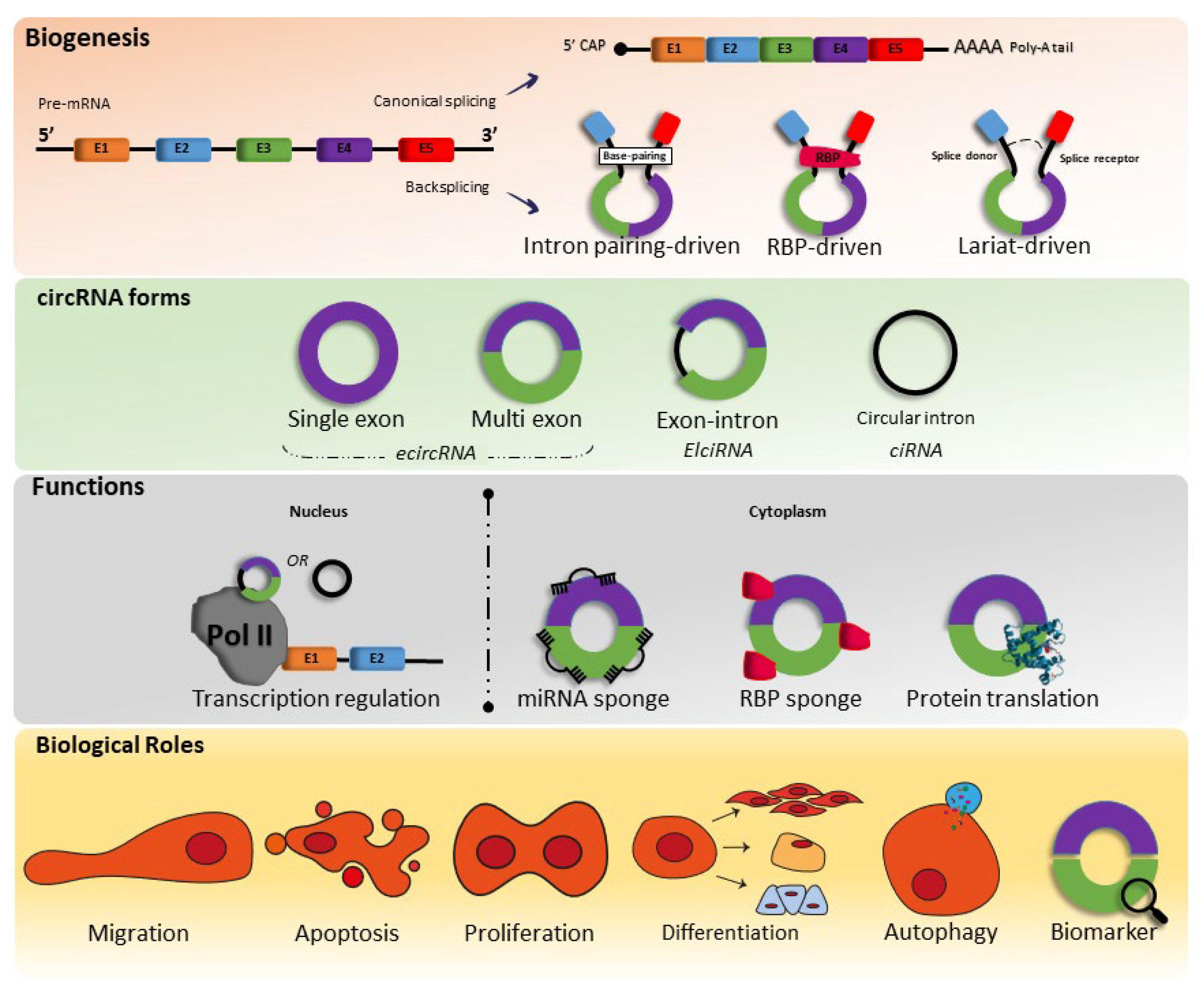
The first circular RNAs (circRNA) molecule that is covalently closed was discovered by Sanger et al., (1976) [1] in plant viroids; since then, some studies associated with circRNAs have been published. However, only in 2010 with advances in RNA sequencing technology did several studies emerge linking circRNAs to diseases such as cardiovascular diseases (CVDs) and cancer [2][3][4][5][6][7][8][9].
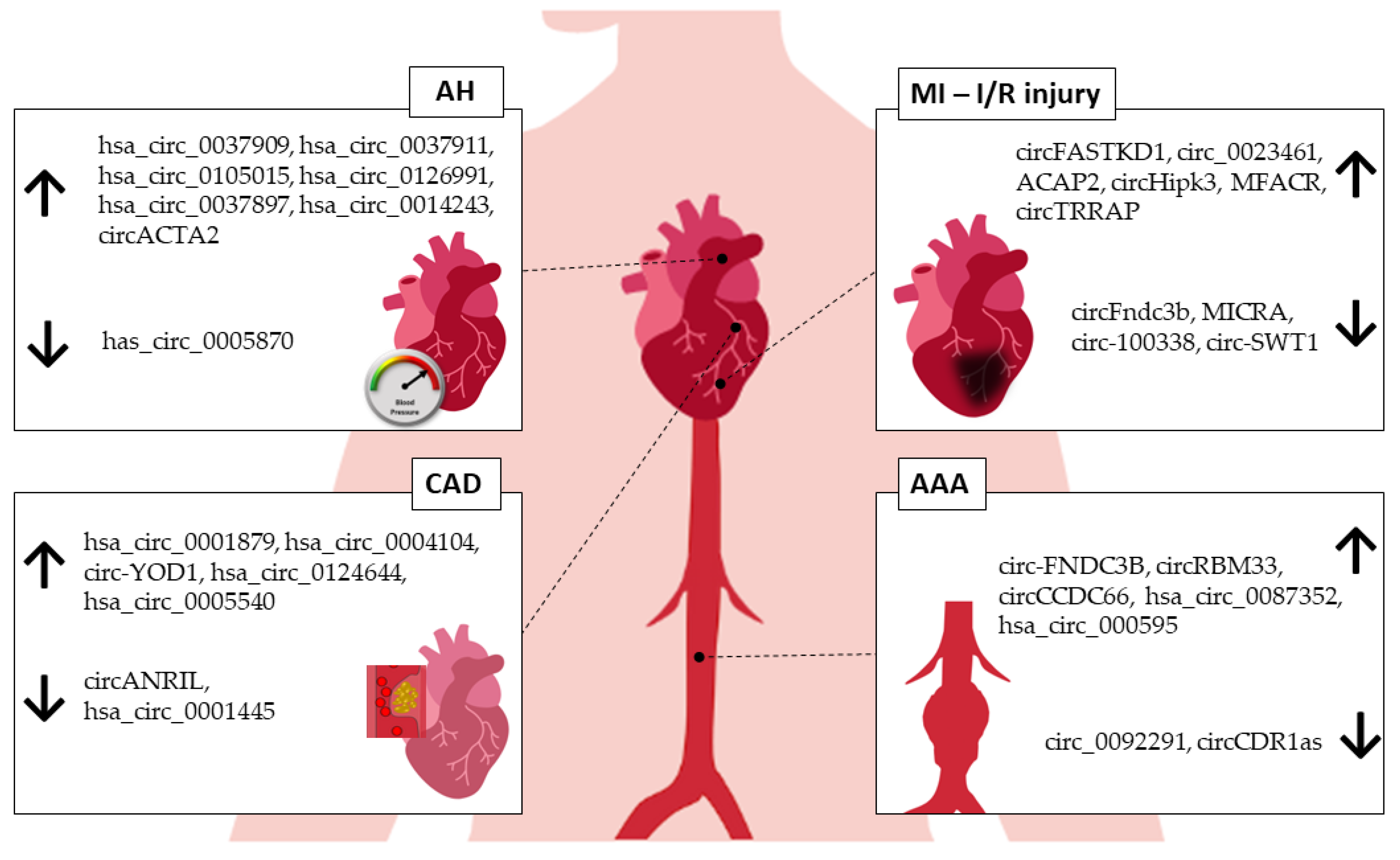
2. circRNAs in Arterial Hypertension
3. circRNAs in Myocardial Infarction
4. circRNAs in Coronary Artery Disease
5. circRNAs in Abdominal Aortic Aneurysm
6. Exercise-Related circRNAs in the Cardiovascular System
References
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856.
- Tan, W.L.W.; Lim, B.T.S.; Anene-Nzelu, C.G.O.; Ackers-Johnson, M.; Dashi, A.; See, K.; Tiang, Z.; Lee, D.P.; Chua, W.W.; Luu, T.D.A.; et al. A Landscape of Circular RNA Expression in the Human Heart. Cardiovasc. Res. 2017, 113, 298–309.
- Werfel, S.; Nothjunge, S.; Schwarzmayr, T.; Strom, T.-M.; Meitinger, T.; Engelhardt, S. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 2016, 98, 103–107.
- Li, Y.; Zhang, J.; Huo, C.; Ding, N.; Li, J.; Xiao, J.; Lin, X.; Cai, B.; Zhang, Y.; Xu, J. Dynamic Organization of lncRNA and Circular RNA Regulators Collectively Controlled Cardiac Differentiation in Humans. eBioMedicine 2017, 24, 137–146.
- Fan, X.; Weng, X.; Zhao, Y.; Chen, W.; Gan, T.; Xu, D. Circular RNAs in Cardiovascular Disease: An Overview. BioMed Res. Int. 2017, 2017, 1–9.
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984.
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.-M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e813.
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.; Zhang, J.-L.; Yang, L.; Chen, L.-L. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016, 15, 611–624.
- Liang, D.; Wilusz, J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014, 28, 2233–2247.
- LiLi, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264.
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular Intronic Long Noncoding RNAs. Mol. Cell 2013, 51, 792–806.
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157.
- Brouwers, S.; Sudano, I.; Kokubo, Y.; Sulaica, E.M. Arterial hypertension. Lancet 2021, 398, 249–261.
- Liu, Y.; Dong, Y.; Dong, Z.; Song, J.; Zhang, Z.; Liang, L.; Liu, X.; Sun, L.; Li, X.; Zhang, M.; et al. Expression Profiles of Circular RNA in Aortic Vascular Tissues of Spontaneously Hypertensive Rats. Front. Cardiovasc. Med. 2021, 8, 814402.
- Ma, Y.; Zheng, B.; Zhang, X.-H.; Nie, Z.-Y.; Yu, J.; Zhang, H.; Wang, D.-D.; Shi, B.; Bai, Y.; Yang, Z.; et al. circACTA2 mediates Ang II-induced VSMC senescence by modulation of the interaction of ILF3 with CDK4 mRNA. Aging 2021, 13, 11610–11628.
- Zhang, J.-R.; Sun, H.-J. LncRNAs and circular RNAs as endothelial cell messengers in hypertension: Mechanism insights and therapeutic potential. Mol. Biol. Rep. 2020, 47, 5535–5547.
- Weiser-Evans, M.C.M. Smooth Muscle Differentiation Control Comes Full Circle. Circ. Res. 2017, 121, 591–593.
- Sun, Y.; Yang, Z.; Zheng, B.; Zhang, X.-H.; Zhang, M.-L.; Zhao, X.-S.; Zhao, H.-Y.; Suzuki, T.; Wen, J.K. A Novel Regulatory Mechanism of Smooth Muscle α-Actin Expression by NRG-1/circACTA2/miR-548f-5p AxisNovelty and Significance. Circ. Res. 2017, 121, 628–635.
- Anderson, J.L.; Morrow, D.A. Acute myocardial infarction. New Engl. J. Med. 2017, 376, 2053–2064.
- Wen, Z.-J.; Xin, H.; Wang, Y.-C.; Liu, H.-W.; Gao, Y.-Y.; Zhang, Y.-F. Emerging roles of circRNAs in the pathological process of myocardial infarction. Mol. Ther. Nucleic Acids 2021, 26, 828–848.
- Gong, C.; Zhou, X.; Lai, S.; Wang, L.; Liu, J. Long Noncoding RNA/Circular RNA-MiRNA-MRNA Axes in Ischemia-Reperfusion Injury. Biomed. Res. Int. 2020, 2020, 8838524.
- Marinescu, M.; Lazar, A.; Marta, M.M.; Cozma, A.; Catana, C. Non-Coding RNAs: Prevention, Diagnosis, and Treatment in Myocardial Ischemia—Reperfusion Injury. Int. J. Mol. Sci. 2022, 23, 2728.
- Huang, F.; Mai, J.; Chen, J.; He, Y.; Chen, X. Non-coding RNAs modulate autophagy in myocardial ischemia-reperfusion injury: A systematic review. J. Cardiothorac. Surg. 2021, 16, 140.
- Zhou, J.; Wang, B.; Bin, X.; Xie, C.; Li, B.; Liu, O.; Tang, Z. CircHIPK3: Key Player in Pathophysiology and Potential Diagnostic and Therapeutic Tool. Front. Med. 2021, 8, 615417.
- Si, X.; Zheng, H.; Wei, G.; Li, M.; Li, W.; Wang, H.; Guo, H.; Sun, J.; Li, C.; Zhong, S.; et al. circRNA Hipk3 Induces Cardiac Regeneration after Myocardial Infarction in Mice by Binding to Notch1 and miR-133a. Mol. Ther. Nucleic Acids 2020, 21, 636–655.
- Wang, Y.; Zhao, R.; Shen, C.; Liu, W.; Yuan, J.; Li, C.; Deng, W.; Wang, Z.; Zhang, W.; Ge, J.; et al. Exosomal CircHIPK3 Released from Hypoxia-Induced Cardiomyocytes Regulates Cardiac Angiogenesis after Myocardial Infarction. Oxidative Med. Cell. Longev. 2020, 2020, 1–19.
- Salgado-Somoza, A.; Zhang, L.; Vausort, M.; Devaux, Y. The circular RNA MICRA for risk stratification after myocardial infarction. IJC Heart Vasc. 2017, 17, 33–36.
- Vausort, M.; Salgado-Somoza, A.; Zhang, L.; Leszek, P.; Scholz, M.; Teren, A.; Burkhardt, R.; Thiery, J.; Wagner, D.R.; Devaux, Y. Myocardial Infarction-Associated Circular RNA Predicting Left Ventricular Dysfunction. J. Am. Coll. Cardiol. 2016, 68, 1247–1248.
- Nowbar, A.N.; Gitto, M.; Howard, J.P.; Francis, D.P.; Al-Lamee, R. Mortality from Ischemic Heart Disease: Analysis of Data from the World Health Organization and Coronary Artery Disease Risk Factors from NCD Risk Factor Collaboration. Circ. Cardiovasc. Qual. Outcomes 2019, 12, e005375.
- Malakar, A.K.; Choudhury, D.; Halder, B.; Paul, P.; Uddin, A.; Chakraborty, S. A review on coronary artery disease, its risk factors, and therapeutics. J. Cell. Physiol. 2019, 234, 16812–16823.
- Cao, Q.; Guo, Z.; Du, S.; Ling, H.; Song, C. Circular RNAs in the pathogenesis of atherosclerosis. Life Sci. 2020, 255, 117837.
- Gong, X.; Tian, M.; Cao, N.; Yang, P.; Xu, Z.; Zheng, S.; Liao, Q.; Chen, C.; Zeng, C.; Jose, P.A.; et al. Circular RNA circEsyt2 regulates vascular smooth muscle cell remodeling via splicing regulation. J. Clin. Investig. 2021, 131, e147031.
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429.
- Wang, L.; Shen, C.; Wang, Y.; Zou, T.; Zhu, H.; Lu, X.; Li, L.; Yang, B.; Chen, J.; Chen, S.; et al. Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis 2019, 286, 88–96.
- Zhao, Z.; Li, X.; Gao, C.; Jian, D.; Hao, P.; Rao, L.; Li, M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 2017, 7, 39918.
- DeRoo, E.; Stranz, A.; Yang, H.; Hsieh, M.; Se, C.; Zhou, T. Endothelial Dysfunction in the Pathogenesis of Abdominal Aortic Aneurysm. Biomolecules 2022, 12, 509.
- Sampson, U.K.A.; Norman, P.E.; Fowkes, F.G.R.; Aboyans, V.; Song, Y.; Harrell, F.E.H., Jr.; Forouzanfar, M.H.; Naghavi, M.; Denenberg, J.O.; McDermott, M.M.; et al. Global and Regional Burden of Aortic Dissection and Aneurysms: Mortality Trends in 21 World Regions, 1990 to 2010. Glob. Heart 2014, 9, 171–180.e10.
- Kessler, V.; Klopf, J.; Eilenberg, W.; Neumayer, C.; Brostjan, C. AAA Revisited: A Comprehensive Review of Risk Factors, Management, and Hallmarks of Pathogenesis. Biomedicines 2022, 10, 94.
- Wang, Z.; You, Y.; Yin, Z.; Bao, Q.; Lei, S.; Yu, J.; Xie, C.; Ye, F.; Xie, X. Burden of Aortic Aneurysm and Its Attributable Risk Factors from 1990 to 2019: An Analysis of the Global Burden of Disease Study 2019. Front. Cardiovasc. Med. 2022, 9, 901225.
- Bath, M.F.; Gokani, V.J.; Sidloff, D.A.; Jones, L.R.; Choke, E.; Sayers, R.D.; Bown, M.J. Systematic review of cardiovascular disease and cardiovascular death in patients with a small abdominal aortic aneurysm. Br. J. Surg. 2015, 102, 866–872.
- Haller, S.J.; Azarbal, A.F.; Rugonyi, S. Predictors of Abdominal Aortic Aneurysm Risks. Bioengineering 2020, 7, 79.
- Miyake, T.; Miyake, T.; Kurashiki, T.; Morishita, R. Molecular Pharmacological Approaches for Treating Abdominal Aortic Aneurysm. Ann. Vasc. Dis. 2019, 12, 137–146.
- Zhou, M.; Shi, Z.; Cai, L.; Li, X.; Ding, Y.; Xie, T.; Fu, W. Circular RNA expression profile and its potential regulative role in human abdominal aortic aneurysm. BMC Cardiovasc. Disord. 2020, 20, 70.
- Kuivaniemi, H.; Ryer, E.J.; Elmore, J.R.; Tromp, G. Understanding the pathogenesis of abdominal aortic aneurysms. Expert Rev. Cardiovasc. Ther. 2015, 13, 975–987.
- Di Gregoli, K.; Anuar, N.N.M.; Bianco, R.; White, S.J.; Newby, A.C.; George, S.J.; Johnson, J.L. MicroRNA-181b Controls Atherosclerosis and Aneurysms Through Regulation of TIMP-3 and Elastin. Circ. Res. 2017, 120, 49–65.
- Sun, X.; He, S.; Wara, A.K.M.; Icli, B.; Shvartz, E.; Tesmenitsky, Y.; Belkin, N.; Li, D.; Blackwell, T.S.; Sukhova, G.K.; et al. Systemic Delivery of MicroRNA-181b Inhibits Nuclear Factor-κB Activation, Vascular Inflammation, and Atherosclerosis in Apolipoprotein E–Deficient Mice. Circ. Res. 2014, 114, 32–40.
- Si, K.; Lu, D.; Tian, J. Integrated analysis and the identification of a circRNA-miRNA-mRNA network in the progression of abdominal aortic aneurysm. PeerJ 2021, 9, e12682.
- Han, Y.; Zhang, H.; Bian, C.; Chen, C.; Tu, S.; Guo, J.; Wu, Y.; Böckler, D.; Zhang, J. Circular RNA Expression: Its Potential Regulation and Function in Abdominal Aortic Aneurysms. Oxidative Med. Cell. Longev. 2021, 2021, 1–21.
- Correia, C.C.M.; Rodrigues, L.F.; Pelozin, B.R.D.A.; Oliveira, E.M.; Fernandes, T. Long Non-Coding RNAs in Cardiovascular Diseases: Potential Function as Biomarkers and Therapeutic Targets of Exercise Training. Non-Coding RNA 2021, 7, 65.
- Wang, L.; Lv, Y.; Li, G.; Xiao, J. MicroRNAs in heart and circulation during physical exercise. J. Sport Health Sci. 2018, 7, 433–441.
- Guo, M.; Qiu, J.; Shen, F.; Wang, S.; Yu, J.; Zuo, H.; Yao, J.; Xu, S.; Hu, T.; Wang, D.; et al. Comprehensive analysis of circular RNA profiles in skeletal muscles of aging mice and after aerobic exercise intervention. Aging 2020, 12, 5071–5090.
- Niu, Y.; Wan, C.; Zhang, J.; Zhang, S.; Zhao, Z.; Zhu, L.; Wang, X.; Ren, X.; Wang, J.; Lei, P. Aerobic exercise improves VCI through circRIMS2/miR-186/BDNF-mediated neuronal apoptosis. Mol. Med. 2021, 27, 4.
- Meinecke, A.; Mitzka, S.; Just, A.; Cushman, S.; Stojanović, S.D.; Xiao, K.; Mooren, F.C.; Fiedler, J.; Thum, T. Cardiac endurance training alters plasma profiles of circular RNA MBOAT2. Am. J. Physiol. Heart Circ. Physiol. 2020, 319, H13–H21.
- Zhu, Y.; Zheng, C.; Zhang, R.; Yan, J.; Li, M.; Ma, S.; Chen, K.; Chen, L.; Liu, J.; Xiu, J.; et al. Circ-Ddx60 contributes to the antihypertrophic memory of exercise hypertrophic preconditioning. J. Adv. Res. 2022, in press.
- Lin, H.; Zhu, Y.; Zheng, C.; Hu, D.; Ma, S.; Chen, L.; Wang, Q.; Chen, Z.; Xie, J.; Yan, Y.; et al. Antihypertrophic Memory After Regression of Exercise-Induced Physiological Myocardial Hypertrophy Is Mediated by the Long Noncoding RNA Mhrt779. Circulation 2021, 143, 2277–2292.





