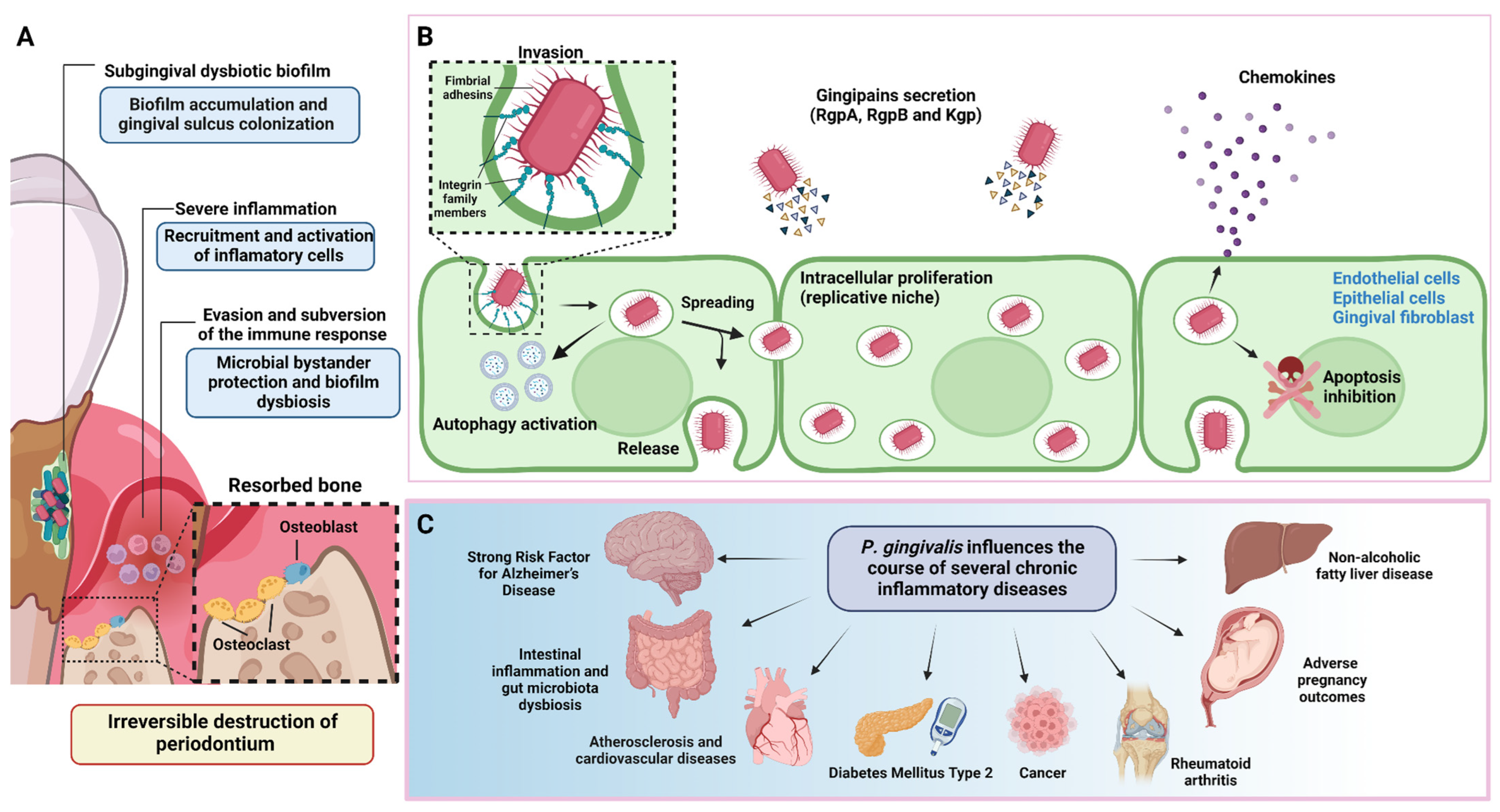
Porphyromonas gingivalis is a well-characterized member of the dysbiotic subgingival microbiota and a key bacterium in the aetiopathogenesis of periodontitis
[6][7]. It is an anaerobe, asaccharolytic, non-motile, Gram-negative bacterium associated with destructive chronic periodontitis
[8]. Its association with periodontitis is based fundamentally on its virulence properties, recurrent presence in affected sites, and higher serum antibody levels in patients diagnosed with chronic periodontitis. In addition, periodontal health indicators are inversely correlated with the presence of this periodontal pathogen
[8][9][10]. Moreover, periodontitis induced by
P. gingivalis may influence the course and pathogenesis of other chronic inflammatory disorders
[11].
P. gingivalis expresses several virulence factors that enable it to colonize, invade, and disrupt periodontal tissues
[8][12]. Despite its low-relative abundance in the subgingival biofilm,
P. gingivalis operates as a keystone pathogen, being critical in promoting the host inflammatory and osteolytic response involved in periodontitis onset and progression
[7][13][14]. Furthermore,
P. gingivalis significantly changes the bacterial taxa that comprise symbiotic microbial communities for heterotypical communities with recognized destructive inflammatory potential
[6][7] (
Figure 1).
P. gingivalis strain virulence varies among different types, and this diversity may alter its keystone pathogen capacity
[14][15][16]. Consequently, precise analysis methods are required for a better understanding of
P. gingivalis in the context of its different pathogenic potential and its modulating effect on multispecies heterotopic communities in the periodontitis-related biofilm.
Figure 1. Periodontitis pathogenesis and P. gingivalis overview. (A) During the onset and progression of periodontitis, P. gingivalis resides in the subgingival biofilm adhered to the tooth surface, where it interacts metabolically with other bacteria, inducing them to express different virulence factors with pathogenic potential. In parallel, P. gingivalis acts as a keystone pathogen, altering the regulation of the immune response in the susceptible host. The metabolic synergism and immune response subversion provide the nutritional and protective conditions required by the dysbiotic subgingival community to increase their diversity and abundance, with the concomitant induction of a strong, destructive inflammatory response. Together, all these activities cause irreversible connective tissue breakdown and resorption of the tooth-supporting alveolar bone, the critical hallmark of periodontitis that causes tooth loss. (B) To invade the periodontium, P. gingivalis uses various virulence factors that allow it to colonize, replicate, and spread in different subsets of cells to increase its progeny and generate infection. (C) In addition to causing tooth loss, P. gingivalis-induced periodontitis can also affect systemic health, influencing the course of other diseases and conditions.
2. P. gingivalis Is a Keystone Pathogen of Periodontitis
As a keystone pathogen,
P. gingivalis disrupts the host’s immune response even at low relative abundances
[22][23]. This microbe generates protection and the optimal nutritional conditions for the growth and development of pathobiont microorganisms, responsible for maintaining the destructive chronic inflammatory environment
[24]. Therefore,
P. gingivalis modulates the entire bacterial community’s composition, abundance, and adaptive fitness
[25][26]. This synergistic interaction was evidenced in a mechanistic study using germ-free mice. In contrast to wild-type mice, the oral microbiota lack in germ-free mice did not induce destructive inflammation after
P. gingivalis inoculation into the periodontal tissues
[13].
The oral microbiome comprises over 600 prevalent taxa distributed in different ecological niches. Because many of these species cannot be cultured, omics technologies are powerful tools for assessing their composition
[27]. Using clinical samples, metagenomics and metatranscriptomics studies have confirmed the prominent subgingival microbial dysbiosis in individuals with periodontitis. Furthermore, they established the presence and relative abundance of
P. gingivalis and the virulence factors upregulation associated with its pathogenesis
[19][28][29][30].
The synergy interactions of the resident microbiota are fundamental in this new periodontitis model. In this sense,
Streptococcus gordonii is an accessory pathogen that contributes to increasing
P. gingivalis virulence
[7]. LuxS is a soluble mediator autoinducer 2 (AI-2) molecule of
P. gingivalis involved in the uptake of hemin/inorganic iron and quorum sensing interspecies with
Streptococcus gordonii. Utilizing transcriptomics was demonstrated in a LuxS-deficient strain that
P. gingivalis fails to form microcolonies, confirming the significance of these molecules in the signaling involved in the formation and maturation of the biofilm
[31]. The role of keystone bacteria has also been evaluated
in vitro. A multispecies symbiotic biofilm (oral health) was generated in the presence or absence of
P. gingivalis and
A. actinomycetemcomitans. The transcriptomic analyses showed significant gene expression pattern differences when comparing both conditions. Similarly, they exhibited different gene expressions when planktonic, and biofilm conditions were compared. These findings showed a two-way modulatory effect of periodontal pathogens on multispecies heterotypic communities
[32]. The keystone pathogen effect has also been evaluated in
in vivo models. The ligature-induced periodontitis model in mice induces microbial dysbiosis and recreates the destructive inflammatory conditions exhibited during periodontitis in humans
[33]. Sterile silk ligature alone causes alveolar bone resorption. However, when it is preincubated with
P. gingivalis and subsequently tied to the tooth, the concomitant bone loss is significantly exacerbated
[34]. Consistent with the systemic effects of periodontitis, oral administration of
P. gingivalis significantly alters the gut microbiome. The metagenomic studies showed evident differences in the composition and relative abundance of the community members compared to mice that did not receive
P. gingivalis. In addition, dysbiosis generates systemic inflammatory effects, changes in the serum metabolome, and aggravates collagen-induced arthritis
[35][36][37][38]. This evidence reveals the contribution of omics disciplines to determining the keystone role of
P. gingivalis in perturbing bacterial communities.
3. Proteomics and Metabolomics in P. gingivalis Pathogenesis
Proteomics corresponds to the study (
Table 1) of the structure, function, and protein interaction on a large scale
[39]. This technique has become a powerful tool for protein identification involved in host-
P. gingivalis interactions and their relationship with other oral bacteria. For example, proteomic studies using mixed oral bacteria cultures showed that nearly 40% of
P. gingivalis proteins can be regulated by the presence of other members of the oral microbiota, such as
Fusobacterium nucleatum,
Streptococcus gordonii, or
Streptococcus oralis [40][41], suggesting an effect of the ecological community on
P. gingivalis protein expression.
Table 1. Summary of metabolomics and proteomics research studies.
The identification and characterization of proteins involved in bacterial colonization and invasion is another relevant scope of
P. gingivalis proteomics. Notable changes have been reported in the extracellular proteomic profiles and virulence factors expressed by different strains of
P. gingivalis [42]. For instance, using mass spectrometry, alterations in the
P. gingivalis O-glycoproteome are evidenced, determining that gingipains lacked these post-translational modifications
[43]. On the other hand, a previously known marker of rheumatoid arthritis is the loss of tolerance to the presence of proteins that have undergone citrullination. This post-translational modification affects arginine residues and produces changes in the structure and function of proteins
[49]. Interestingly,
P. gingivalis contains a peptidyl-arginine deiminase (PPAD), an enzyme capable of protein citrullination, and it can modify bacterial and human proteins
[42]. Moreover, the comparison of secreted protein citrullination profiles among several
P. gingivalis clinical isolates with reference strains showed substantial differences, primarily focused on six to 25 proteins, mostly composed of virulence factors. The role of these modifications remains to be clarified.
Proteomics can also help to understand the role of some cellular derivates, such as Outer membrane vesicles (OMVs).
P. gingivalis releases several virulence factors toward the environment via OMVs
[44]. Studies covering the role of OMV-associated proteome in
P. gingivalis suggest important changes according to the availability of heme, displaying an upregulation of proteins involved in heme binding and transport when this nutrient is limited
[45]. Collectively, proteomic studies reveal that the
P. gingivalis proteome changes depending on the interaction target and the biological context, offering another opportunity to understand
P. gingivalis physiology and pathogenesis.
Another useful technique to understand the role of
P. gingivalis in periodontitis is metabolomics (
Table 1), which is the systematic study of metabolic content produced and consumed by specific cellular processes. Thus, it presents the collection of all metabolites within a particular sample
[50]. Current evidence suggests that during periodontitis, a metabolic alteration in the oral cavity is induced by the colonization of pathogenic bacteria
[48]. Additionally, metabolic changes are also reported in cells of the periodontal ligament (PDLSCs) under
P. gingivalis infection, showing a metabolic reprogramming that compromises processes associated with the glycolysis, tricarboxylic acid (TCA) cycle, tryptophan, and choline metabolism
[47][48]. Since gene sequence and expression profiles are essential but not sufficient to understand the mechanisms underlying
P. gingivalis pathogenesis, proteomic and metabolomic studies have begun to gain more insights into periodontitis-related host-pathogen interactions.