Periodontitis is a non-communicable chronic inflammatory disease characterized by the progressive and irreversible breakdown of the soft periodontal tissues and resorption of teeth-supporting alveolar bone. The etiology of periodontitis involves dysbiotic shifts in the diversity of microbial communities inhabiting the subgingival crevice, which is dominated by anaerobic Gram-negative bacteria, including
Porphyromonas gingivalis
. Indeed,
P. gingivalis
is a keystone pathogen with a repertoire of attributes that allow it to colonize periodontal tissues and influence the metabolism, growth rate, and virulence of other periodontal bacteria. The pathogenic potential of
P. gingivalis
has been traditionally analyzed using classical biochemical and molecular approaches.
- periodontitis
- microbial dysbiosis
- Porphyromonas gingivalis
- keystone pathogen
1. Introduction
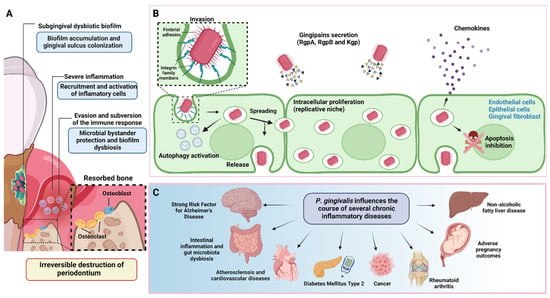
2. P. gingivalis Is a Keystone Pathogen of Periodontitis
As a keystone pathogen, P. gingivalis disrupts the host’s immune response even at low relative abundances [22][23][60,61]. This microbe generates protection and the optimal nutritional conditions for the growth and development of pathobiont microorganisms, responsible for maintaining the destructive chronic inflammatory environment [24][62]. Therefore, P. gingivalis modulates the entire bacterial community’s composition, abundance, and adaptive fitness [25][26][63,64]. This synergistic interaction was evidenced in a mechanistic study using germ-free mice. In contrast to wild-type mice, the oral microbiota lack in germ-free mice did not induce destructive inflammation after P. gingivalis inoculation into the periodontal tissues [13]. The oral microbiome comprises over 600 prevalent taxa distributed in different ecological niches. Because many of these species cannot be cultured, omics technologies are powerful tools for assessing their composition [27][65]. Using clinical samples, metagenomics and metatranscriptomics studies have confirmed the prominent subgingival microbial dysbiosis in individuals with periodontitis. Furthermore, they established the presence and relative abundance of P. gingivalis and the virulence factors upregulation associated with its pathogenesis [19][28][29][30][19,34,46,66]. The synergy interactions of the resident microbiota are fundamental in this new periodontitis model. In this sense, Streptococcus gordonii is an accessory pathogen that contributes to increasing P. gingivalis virulence [7]. LuxS is a soluble mediator autoinducer 2 (AI-2) molecule of P. gingivalis involved in the uptake of hemin/inorganic iron and quorum sensing interspecies with Streptococcus gordonii. Utilizing transcriptomics was demonstrated in a LuxS-deficient strain that P. gingivalis fails to form microcolonies, confirming the significance of these molecules in the signaling involved in the formation and maturation of the biofilm [31][55]. The role of keystone bacteria has also been evaluated in vitro. A multispecies symbiotic biofilm (oral health) was generated in the presence or absence of P. gingivalis and A. actinomycetemcomitans. The transcriptomic analyses showed significant gene expression pattern differences when comparing both conditions. Similarly, they exhibited different gene expressions when planktonic, and biofilm conditions were compared. These findings showed a two-way modulatory effect of periodontal pathogens on multispecies heterotypic communities [32][57]. The keystone pathogen effect has also been evaluated in in vivo models. The ligature-induced periodontitis model in mice induces microbial dysbiosis and recreates the destructive inflammatory conditions exhibited during periodontitis in humans [33][67]. Sterile silk ligature alone causes alveolar bone resorption. However, when it is preincubated with P. gingivalis and subsequently tied to the tooth, the concomitant bone loss is significantly exacerbated [34][68]. Consistent with the systemic effects of periodontitis, oral administration of P. gingivalis significantly alters the gut microbiome. The metagenomic studies showed evident differences in the composition and relative abundance of the community members compared to mice that did not receive P. gingivalis. In addition, dysbiosis generates systemic inflammatory effects, changes in the serum metabolome, and aggravates collagen-induced arthritis [35][36][37][38][69,70,71,72]. This evidence reveals the contribution of omics disciplines to determining the keystone role of P. gingivalis in perturbing bacterial communities.3. Proteomics and Metabolomics in P. gingivalis Pathogenesis
Proteomics corresponds to the study (Table 1) of the structure, function, and protein interaction on a large scale [39][73]. This technique has become a powerful tool for protein identification involved in host-P. gingivalis interactions and their relationship with other oral bacteria. For example, proteomic studies using mixed oral bacteria cultures showed that nearly 40% of P. gingivalis proteins can be regulated by the presence of other members of the oral microbiota, such as Fusobacterium nucleatum, Streptococcus gordonii, or Streptococcus oralis [40][41][74,75], suggesting an effect of the ecological community on P. gingivalis protein expression.| Strategy | Method and Sample | Findings/Contributions | Reference | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proteomics | Cultured | P. gingivalis | , | F. nucleatum | , and | S. gordonii | mixed biofilm and | P. gingivalis | monobiofilm as control. Bacterial cells were lysed, and proteins were digested for mass spectrometry. | The proteome of the mixed biofilm differs from the monobiofilm, it exhibits a decrease in proteins involved in cell shape and cell envelope formation, and an increase in HmuR protein (an outer membrane receptor). | [40] | [74] | ||||||||
| Proteomics | Cultured | P. gingivalis | (ATCC | ® | 33277™) and | S. oralis | (ATCC | ® | 9811™) mixed biofilm. Controls are monobiofilm of | P. gingivalis | and | S. oralis | . Biofilm samples were digested and then summited to shotgun proteomic analysis with LC-MS/MS. | The | P. gingivalis | proteins that increased their expression induced by the interaction with | S. oralis | were GyrB, RpoD, FimA and a probable transcriptional regulatory protein. | [41] | [75] |
| Proteomics | Liquid cultured | P. gingivalis | (ATCC | ® | 33277™, W83 and two peptidylarginine deiminase (PPAD) mutant strains) were centrifugated and the supernatant was analyzed by mass spectrometry analysis. | Analysis of the | P. gingivalis | proteome and extracellular citrulilnome showed heterogeneity between the different isolates. Furthermore, the main virulence factors revealed different patterns in their citrullination. | [42] | [76] | ||||||||||
| Proteomics | Cultured | P. gingivalis | ATCC | ® | 33277™ and mutant strains. Cells were harvested, lysed, and the supernatant was subjected to mass spectrometry analysis. | Identification of 257 putative O-glycosylation sites within 145 glycoproteins of | P. gingivalis | . Demonstration for the first time the presence of the O-glycosylation system in | P. gingivalis | . | [43] | [77] | ||||||||
| Proteomics | Cultured | P. gingivalis | (W50 strain), then cells were harvested and centrifuged, and the supernatant was filtered to obtain outer membrane vesicles (OMVs) for mass spectrometry analysis. | A total of 151 OMV proteins were identified and the most enriched proteins were LptO, IhtB and HmuY. | [44] | [78] | ||||||||||||||
| Proteomics | Cultured | P. gingivalis | (W50 strain) in three conditions: control, heme limitation, and heme excess conditions. Then cells were harvested and processed to obtain whole cell lysate and outer membrane vesicles separately and then mass spectrometry analyses. | The proteins most upregulated in response to heme limitation were those involved in binding and transporting heme. | [45] | [79] | ||||||||||||||
| Metabolomics | Tongue swabs and mouth washout samples from patients with chronic periodontal disease were analyzed with proton nuclear magnetic resonance (H-NMR) to determine their metabolic status. | The metabolic state of the mouth of chronic periodontal disease patients changes in the levels of eight metabolites in comparison to healthy individuals. These metabolic changes could be used as a periodontal disease-associated process biomarker. | [46] | [80] | ||||||||||||||||
| Metabolomics | Meditation through Gas chromatography-mass spectrometer (GC-MS) metabolite profiling of cultured human periodontal ligament fibroblast infected with | P. gingivalis | (ATCC | ® | 33277™). | Periodontal ligament cells (PDLSCs) experienced metabolic reprogramming due to the infection of | P. gingivalis | . These metabolic changes could be related to pro-inflammatory responses on PDLSC, showing a shift from oxidative phosphorylation to glycolysis. | [47] | [81] | ||||||||||
| Metabolomics and metagenomics | Serum samples from mice submitted to an oral gavage of | P. gingivalis | (ATCC | ® | 33277 | TM | ) and sham control were analyzed with Untargeted metabolomics profiling chromatographic separation and mass spectrometry (MS). Additionally, RNA extraction and metagenomic analysis were done in colon samples from the same study groups. | The analysis of the metabolites in | P. gingivalis-administered | mice demonstrated that oral administration of this periodontal pathogen could induce dysbiosis of the gut microbiota. In addition, these derived metabolites are associated with metabolic pathways and could be related to the development of metabolic disorders and the destruction of intestinal barrier function. | [48] | [82] |
