
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Prasanta Kumar Choudhury | -- | 1258 | 2022-12-13 05:45:33 | | | |
| 2 | Conner Chen | -9 word(s) | 1249 | 2022-12-14 08:55:17 | | | | |
| 3 | Conner Chen | + 2 word(s) | 1251 | 2022-12-14 08:55:56 | | |
Video Upload Options
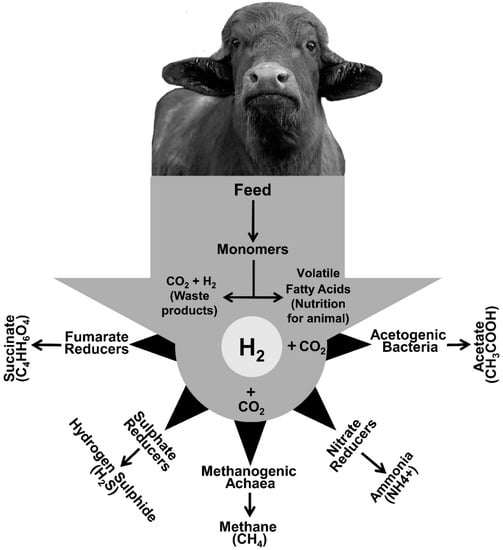
Climate change and the urgent need to reduce greenhouse gas (GHG) emission from agriculture has resulted in significant pressure on the livestock industry for advanced practices that are environmentally more sustainable. Livestock is responsible for more than 15% of anthropogenic methane (CH4) emission via enteric fermentation and improved strategies for mitigating enteric CH4 production therefore represents a promising target to reduce the overall GHG contribution from agriculture. Ruminal CH4 is produced by methanogenic archaea, combining carbon dioxide (CO2) and hydrogen (H2). Removal of H2 is essential, as its accumulation inhibits many biological functions that are essential for maintaining a healthy rumen ecosystem.
1. Introduction
2. Hydrogen: A Key Player in Rumen Fermentation
References
- Wang, S.; Giller, K.; Kreuzer, M.; Ulbrich, S.E.; Braun, U.; Schwarm, A. Contribution of ruminal fungi, archaea, protozoa, and bacteria to the methane suppression caused by oilseed supplemented diets. Front. Microbiol. 2017, 8, 1864.
- Choudhury, P.K.; Salem, A.Z.M.; Jena, R.; Kumar, S.; Singh, R.; Puniya, A.K. Rumen microbiology: An overview. In Rumen Microbiology-Evolution to Revolution; Puniya, A.K., Singh, R., Kamra, D.N., Eds.; CRC Springer: New Delhi, India, 2015; pp. 3–16.
- Kumar, S.; Dagar, S.S.; Puniya, A.K.; Upadhyay, R.C. Changes in methane emission, rumen fermentation in response to diet and microbial interactions. Res. Vet. Sci. 2013, 94, 263–268.
- Kamra, D.N. Rumen microbial ecosystem. Curr. Sci. 2005, 89, 124–135.
- Janssen, P.H.; Kirs, M. Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 2008, 74, 3619–3625.
- Kumar, S.; Choudhury, P.K.; Carro, M.D.; Griffith, G.W.; Dagar, S.S.; Puniya, M.; Calabro, S.; Ravella, S.R.; Dhewa, T.; Upadhyay, R.C.; et al. New aspects and strategies for methane mitigation from ruminants. Appl. Microbiol. Biotechnol. 2014, 98, 31–44.
- Ragsdale, S.W.; Pierce, E. Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim. Biophys. Acta 2008, 1784, 1873–1898.
- Ishaq, S.L.; AlZahal, O.; Walker, N.; McBride, B. An investigation into rumen fungal and protozoal diversity in three rumen fractions, during high-fiber or grain-induced sub-acute ruminal acidosis conditions, with or without active dry yeast supplementation. Front. Microbiol. 2017, 8, 1943.
- Malik, P.K.; Bhatta, R.; Gagen, E.J.; Sejian, V.; Soren, N.M.; Prasad, C.S. Alternate H2 sinks for reducing rumen methanogenesis. In Climate Change Impact on Livestock: Adaptation and Mitigation; Sejian, V., Gaughan, J., Baumgard, L., Prasad, C., Eds.; Springer: New Delhi, India, 2015; pp. 303–320.
- Joblin, K. Methanogenic archaea. In Methods in Gut Microbial Ecology for Ruminants; Makker, H., McSweeney, C.S., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 47–53.
- Moss, A.R.; Jouany, J.P.; Newbold, J. Methane production by ruminants: Its contribution to global warming. Ann. Zootech. 2000, 49, 231–253.
- Ungerfeld, E.M. Inhibition of rumen methanogenesis and ruminant productivity: A meta-analysis. Front. Vet. Sci. 2018, 5, 113.
- Beauchemin, K.A.; Ungerfeld, E.M.; Eckard, R.J.; Wang, M. Review: Fifty years of research on rumen methanogenesis: Lessons learned and future challenges for mitigation. Animal 2020, 14, s2–s16.
- Van Nevel, C.J.; Demeyer, D.I. Control of rumen methanogenesis. Environ. Monit. Assess. 1996, 42, 73–97.
- Demeyer, D.; Fiedler, D.; De Graeve, K.G. Attempted induction of acetogenesis into the rumen fermentation in vitro. Reprod. Nutr. Dev. 1996, 36, 233–240.
- Garsa, A.K.; Choudhury, P.K.; Puniya, A.K.; Dhewa, T.; Malik, R.K.; Tomar, S.K. Bovicins: The bacteriocins of streptococci and their potential in methane mitigation. Probiotics Antimicrob. Proteins 2019, 11, 1403–1413.
- Goopy, J.P. Creating a low enteric methane emission ruminant: What is the evidence of success to the present and prospects for developing economies? Anim. Prod. Sci. 2019, 59, 1769–1776.
- Hook, S.E.; Wright, A.D.G.; McBride, B.W. Methanogens: Methane producers of the rumen and mitigation strategies. Archaea 2010, 2010, 945785.
- Kim, S.H.; Mamuad, L.L.; Islam, M.; Lee, S.S. Reductive acetogens isolated from ruminants and their effect on in vitro methane mitigation and milk performance in Holstein cows. J. Anim. Sci. Technol. 2020, 62, 1–13.
- Martin, C.; Morgavi, D.P.; Doreau, M. Methane mitigation in ruminants: From microbe to the farm scale. Animal 2010, 4, 351–365.
- Wanapat, M.; Kongmun, P.; Poungchompu, O.; Cherdthong, A.; Khejornsart, P.; Pilajun, R.; Kaenpakdee, S. Effects of plants containing secondary compounds and plant oils on rumen fermentation and ecology. Trop. Anim. Health Prod. 2012, 44, 399–405.
- Beauchemin, K.A.; Kreuzer, M.O.; O’Mara, F.P.; McAllister, T.A. Nutritional management for enteric methane abatement: A review. Aust. J. Exp. Agric. 2008, 48, 21–27.
- García-González, R.; López, S.; Fernández, M.; Bodas, R.; González, J.S. Screening the activity of plants and spices for decreasing ruminal methane production in vitro. Anim. Feed Sci. Technol. 2008, 147, 36–52.
- Eckard, R.J.; Grainger, C.; de Klein, C.A.M. Options for the abatement of methane and nitrous oxide from ruminant production: A review. Livest. Sci. 2011, 130, 47–56.
- Patra, A.; Park, T.; Kim, M.; Yu, Z. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J. Anim. Sci. Biotechnol. 2017, 8, 13.
- McGinn, S.M.; Beauchemin, K.A.; Coates, T.; Colombatto, D. Methane emissions from beef cattle: Effects of monensin, sunflower oil, enzymes, yeast and furmaric acid. J. Anim. Sci. 2004, 82, 3346–3356.
- Janssen, P.H.; Frenzel, P. Inhibition of methanogenesis by methyl fluoride: Studies of pure and defined mixed cultures of anaerobic bacteria and archaea. Appl. Environ. Microbiol. 1997, 63, 4552–4557.
- Miller, T.L.; Wolin, M.J. Inhibition of growth of methane-producing bacteria of the ruminant forestomach by hydroxymethylglutaryl-SCoA reductase inhibitors. J. Dairy Sci. 2001, 84, 1445–1448.
- Mitsumori, M.; Shinkai, T.; Takenaka, A.; Enishi, O.; Higuchi, K.; Kobayashi, Y.; Nonaka, I.; Asanuma, N.; Denman, S.E.; McSweeney, C.S. Responses in digestion, rumen fermentation and microbial populations to inhibition of methane formation by a halogenated methane analogue. Br. J. Nutr. 2012, 108, 482–491.
- Sanchez, J.M.; Valle, L.; Rodriguez, F.; Morinnigo, M.A.; Borrego, J.J. Inhibition of methanogenesis by several heavy metals using pure cultures. Lett. Appl. Microbiol. 1996, 23, 439–444.
- Fievez, V.F.; Dohme, M.; Daneels, K.R.; Demeyer, D. Fish oils as potent rumen methane inhibitors and associated effects on rumen fermentation in vitro and in vivo. Anim. Feed Sci. Technol. 2003, 104, 41–58.
- Machmüller, A.; Kreuzer, M. Methane suppression by coconut oil and associated effects on nutrient and energy balance in sheep. Can. J. Anim. Sci. 1999, 79, 65–72.
- Machmüller, A. Medium-chain fatty acids and their potential to reduce methanogenesis in domestic ruminants. Agric. Ecosyst. Environ. 2006, 112, 107–114.
- Lee, S.S.; Hsu, J.T.; Mantovani, H.C.; Russell, J.B. The effect of bovicin HC5, a bacteriocin from Streptococcus bovis HC5, on ruminal methane production in vitro. FEMS Microbiol. Lett. 2002, 217, 51–55.
- Baker, S.K. Rumen methanogens and inhibition of methanogenesis. Aust. J. Agric. Res. 1999, 50, 1293–1298.
- Anderson, R.C.; Carstens, G.E.; Miller, R.K.; Callaway, T.R.; Schultz, C.L.; Edrington, T.S.; Harvey, R.; Nisbet, D. Effect of oral nitroethane and 2-nitropropanol administration on methane-producing activity and volatile fatty acid production in the ovine rumen. Bioresour. Technol. 2006, 97, 2421–2426.
- Wright, A.D.G.; Kennedy, P.; O’neill, C.J.; Toovey, A.F.; Popovski, S.; Rea, S.M.; Pimm, C.L.; Klein, L. Reducing methane emissions in sheep by immunization against rumen methanogens. Vaccine 2004, 22, 3976–3985.
- Zhang, L.; Huang, X.; Xue, B.; Peng, Q.; Wang, Z.; Yan, T.; Wang, L. Immunization against rumen methanogenesis by vaccination with a new recombinant protein. PLoS ONE 2015, 10, e0140086.
- Newbold, C.J.; de la Fuente, G.; Belanche, A.; Ramos-Morales, E.; McEwan, N.R. The Role of Ciliate Protozoa in the Rumen. Front. Microbiol. 2015, 6, 1313.
- LeVan, T.D.; Robinson, J.A.; John, R.; Greening, R.C.; Smolenski, W.J.; Leedle, J.A.Z.; Schaefer, D.M. Assessment of reductive acetogenesis with indigenous ruminal bacterium populations and Acetitomaculum ruminis. Appl. Environ. Microbiol. 1998, 64, 3429–3436.
- Nollet, L.; Demeyer, D.; Verstraete, W. Effect of 2-bromoethanesulfonic acid and Peptostreptococcus productus ATCC35244 addition on stimulation of reductive acetogenesis in the ruminal ecosystem by selective inhibition of methanogenesis. Appl. Environ. Microbiol. 1997, 63, 194–200.
- Nollet, L.; Velde, I.V.; Verstraete, W. Effect of the addition of Peptostreptococcus productus ATCC35244 on the gastro-intestinal microbiota and its activity, as simulated in an in vitro simulator of the human gastro-intestinal tract. Appl. Microbiol. Biotechnol. 1997, 48, 99–104.
- Liu, H.; Wang, J.; Wang, A.; Chen, J. Chemical inhibitors of methanogenesis and putative applications. Appl. Microbiol. Biotechnol. 2011, 89, 1333–1340.
- Hungate, R.E. The Rumen and Its Microbes; Academic Press: New York, NY, USA, 1966; p. 533.
- Ungerfeld, E.M. Metabolic hydrogen flows in rumen fermentation: Principles and possibilities of interventions. Front. Microbiol. 2020, 11, 589.
- Williams, A.G.; Coleman, G.S. The rumen protozoa. In The Rumen Microbial Ecosystem, 2nd ed.; Hobson, P.N., Stewarteds, C.S., Eds.; Blackie Academic & Professional: New York, NY, USA, 1997; pp. 73–139.
- Cord-Ruwisch, R.; Seitz, H.J.; Conrad, R. The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the electron acceptor. Arch. Microbiol. 1988, 149, 350–357.
- Immig, I. The rumen and hindgut as a source of ruminant methanogenesis. Environ. Monit. Assess. 1996, 42, 57–72.
- McAllister, T.A.; Newbold, C.J. Redirecting rumen fermentation to reduce methanogenesis. Aust. J. Exp. Agric. 2008, 48, 7–13.
- Ellis, J.L.; Dijkstra, J.; Kebreab, E.; Bannink, A.; Odongo, N.E.; Mcbride, B.W.; France, J. Aspects of rumen microbiology central to mechanistic modelling of methane production in cattle. J. Agric. Sci. 2008, 146, 213–233.
- Hegarty, R.S.; Gerdes, R. Hydrogen production and transfer in the rumen. Rec. Adv. Anim. Nutr. 1999, 12, 37–44. Available online: http://livestocklibrary.com.au/handle/1234/19891 (accessed on 25 October 2022).
- Greening, C.; Geier, R.; Wang, C.; Woods, L.C.; Morales, S.E.; McDonald, M.J.; Rushton-Green, R.; Morgan, X.C.; Koike, S.; Leahy, S.C.; et al. Diverse hydrogen production and consumption pathways influence methane production in ruminants. ISME J. 2019, 13, 2617–2632.
- Morgavi, D.P.; Forano, E.; Martin, C.; Newbold, C.J. Microbial ecosystem and methanogenesis in ruminants. Animal 2010, 4, 1024–1036.
- Gagen, E.J.; Denman, S.E.; McSweeney, C.S. Acetogenesis as an alternative to methanogenesis in the rumen. In Livestock Production and Climate Change; Malik, P.K., Bhatta, R., Takahashi, J., Kohn, R.A., Prasad, C.S., Eds.; CABI: Wallingford, UK, 2015; pp. 292–303.
- Morvan, B.; Bonnemoy, F.; Fonty, G.; Gouet, P. Quantitative determination of H2-utilizing acetogenic and sulfate-reducing bacteria and methanogenic archaea from digestive tract of different mammals. Curr. Microbiol. 1996, 32, 129–133.
- Kim, S.H.; Mamuad, L.L.; Kim, D.W.; Kim, S.K.; Lee, S.S. Fumarate reductase-producing enterococci reduce methane production in rumen fermentation in vitro. J. Microbiol. Biotechnol. 2016, 26, 558–566.
- Mamuad, L.L.; Kim, S.H.; Lee, S.S.; Cho, K.K.; Jeon, C.O.; Lee, S.S. Characterization, metabolites and gas formation of fumarate reducing bacteria isolated from Korean native goat (Capra hircus coreanae). J. Microbiol. 2012, 50, 925–931.
- Wolin, M.J.; Miller, T.L.; Stewart, C.S. Microbe-microbe interactions. In The Rumen Microbial Ecosystem; Hobson, P.N., Stewart, C.S., Eds.; Chapman and Hall: London, UK, 1997; pp. 467–488.




