Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jianbo Feng | -- | 2623 | 2022-12-06 07:37:47 | | | |
| 2 | Camila Xu | Meta information modification | 2623 | 2022-12-06 08:45:34 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Luo, Y.; Xiang, S.; Feng, J. Protein Phase Separation. Encyclopedia. Available online: https://encyclopedia.pub/entry/38097 (accessed on 09 March 2026).
Luo Y, Xiang S, Feng J. Protein Phase Separation. Encyclopedia. Available at: https://encyclopedia.pub/entry/38097. Accessed March 09, 2026.
Luo, Yan, Shasha Xiang, Jianbo Feng. "Protein Phase Separation" Encyclopedia, https://encyclopedia.pub/entry/38097 (accessed March 09, 2026).
Luo, Y., Xiang, S., & Feng, J. (2022, December 06). Protein Phase Separation. In Encyclopedia. https://encyclopedia.pub/entry/38097
Luo, Yan, et al. "Protein Phase Separation." Encyclopedia. Web. 06 December, 2022.
Copy Citation
Phase separation is a process by which a well-mixed solution of macromolecules such as proteins or nucleic acids spontaneously separates into two phases: a dense phase and a dilute phase.
phase separation
membraneless organelle
biomolecular condensates
1. Introduction
Biological evolution is the evolutionary process of the development of all life forms, and an important characteristic is the progressive complexity and refinement of the different levels of morphological structures. Typically, different biological reactions take place in different organelles in an orderly manner; for example, transcription occurs primarily in the nucleus, protein modification occurs in the endoplasmic reticulum and Golgi, and molecular degradation occurs in lysosomes. These organelles are surrounded by a single or double layer of molecular membranes and are isolated from the surrounding environment. To ensure that various cellular components can aggregate at the correct time and in the proper space to perform their corresponding functions, cellular molecules are isolated in different cellular compartments as needed. In addition to classic membrane-bound organelles, much evidence further suggests that cells possess various membraneless compartments, including the nucleolus, Cajal body, and PcG body in the cellular nucleus [1], along with stress granules (SGs) and P-bodies in the cytoplasm [2][3][4][5]. Proteins, RNA, and other molecular components constitute these membraneless organelles, and phase separation drives the processes involved [2][6]. Although not covered by cell membranes, membraneless organelles are still capable of frequent molecular exchange with the environment. With ongoing scientific inquiry, many insights have been gained into the structure of these organelles. However, questions about why these organelles form, the mechanisms involved, and how their biological properties affect their function have not been answered. As science has advanced, these questions have begun to be resolved, and a deeper understanding of the organization, molecular properties, and regulation of membraneless organelles has emerged [7][8]. In recent years, there has been growing evidence that membraneless organelles are involved in the development of multiple cancers [9][10]. These findings have given rise to a new domain of cellular biology. The emphasis is on learning how cell substances are organized into membraneless organelles, how to promote their activity, and how disorders in these organelles frequently lead to diseases, encouraging researchers to consider the biological processes involved from the viewpoint of phase separation.
Cancer is a disease that seriously threatens human health. Both in terms of morbidity and mortality, cancer is a major global public health problem and second only to cardiovascular disease in terms of mortality [11]. Although research on the occurrence and development of cancer has a history spanning many years, its treatment is still a great problem facing the world. To overcome this challenge, new concepts are urgently needed to characterize and explain the complicated mechanisms of human cancer. An increasing body of evidence suggests that there is a close link between abnormal phase separation condensation assembly and aberrant oncogenic procedures. The advent of protein phase separation offers a novel possibility for targeting refractory cancer.
2. Overview of Phase Separation
Phase separation is a process by which a well-mixed solution of macromolecules such as proteins or nucleic acids spontaneously separates into two phases: a dense phase and a dilute phase [12]. Whether phase separation occurs in a solution depends greatly on the concentration and properties of the macromolecules and the solution, as well as on the environmental conditions [13]. With a growing understanding of the basic molecular principles of phase separation, there is an awareness of the different functions of phase separation in various cellular processes. In general, the main ones include stress response, regulation of gene expression and control of signal transduction [6][14], protein degradation [15], cytoskeleton assembly [16], and gene activation [17] or repression [18], including epigenetics, transcription, and translation. Furthermore, it has been demonstrated that many fundamental biological processes are inseparable from phase separation, including heterochromatin formation [18][19], nucleocytoplasmic transport [20], supramolecular assemblies [21], and assembly of membraneless compartments, such as SGs [22]. Cell architectures formed by phase separation are named biomolecular condensates to mirror their provenance through condensation reactions [2][23]. Contrary to other types of components, they can enrich molecules, and rapid exchange of components and agglomeration of droplets can form specific cellular structures called membraneless organelles by phase separation. In different physiopathological situations, biomolecular condensates can be converted into different states of matter. Similarly, condensates play an essential role in the life activities of various organisms, including advanced structures, gene expression regulation [24], autophagic degradation of incorrectly folded or unneeded proteins [25], signal cluster assembly, and synaptic plasticity regulation of the formation of signaling molecules [10].
2.1. Phase Separation to Form Membraneless Organelles
In 2009, Hyman and Brangwynne found that some properties of P granules resemble liquids and that regulated dissolution/condensation drives their localization, and the researchers first realized that membraneless organelles may be driven by phase separation [26]. Membraneless organelles are dynamic structures with liquid-like physical properties [26]. Due to a lack of lipid-rich membranes, changes in the surrounding environment can easily affect their internal homeostasis, so the protein composition and morphology of membraneless organelles respond accordingly to changes in the cellular environment, and this ability may represent the mechanism underlying the stress response sensed by membraneless organelles [27]. For example, oncogenic ARF protein is localized in the nucleolus and released into the nucleoplasm through changes in phase separation in response to environmental stresses of DNA damage and oncogene activation, hence activating the p53 oncogenic pathway [28].
What is the unique role of membraneless organelles driven by phase separation? The nucleolus is the largest and most intensively studied membraneless organelle, serving as the center of ribosomal RNA synthesis and nascent ribosome assembly in eukaryotic cells. Ribosome biogenesis is vectorial, starting from fibrillar centers (FCs), where rDNA is transcribed into rRNA [29]. Paraspeckles are located in chromatin gaps and are subnuclear bodies built on NEAT1 (a long noncoding RNA). Paraspeckles are involved in many physiological processes, including the cellular stress response, cell differentiation, corpus luteum formation, and cancer development [30]. The proteins that comprise paraspeckles are related to RNA polymerase II transcription and RNA processing. Cajal bodies (CBs) and histone locus bodies (HLBs) could be responsive to stress conditions, and they are nuclear bodies (NBs) involved in the transcriptional and posttranscriptional regulation of small nuclear RNAs and histone genes [31]. With the deepening of research, an increasing number of membraneless organelles and their functions are being discovered (Figure 1).
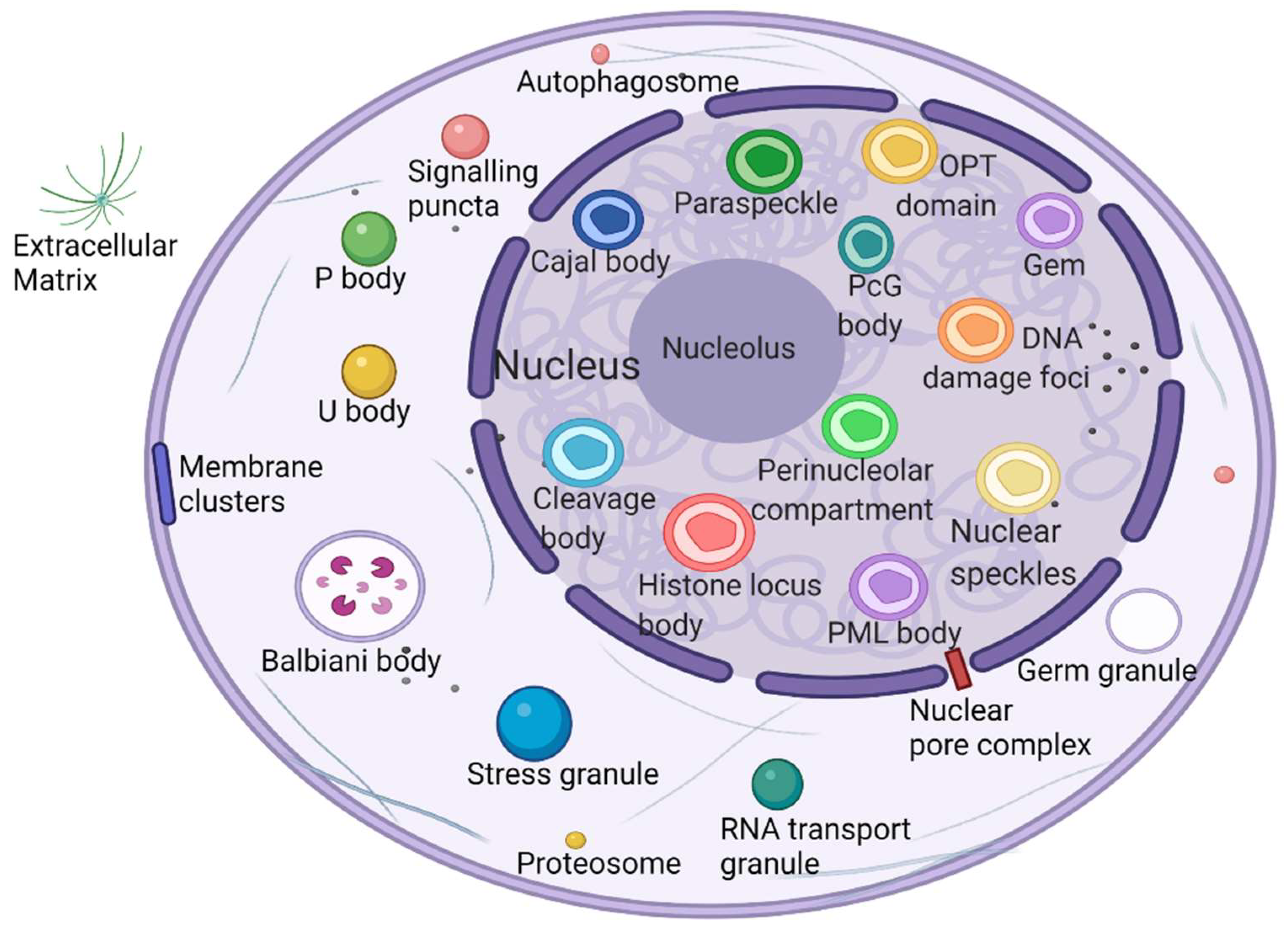
Figure 1. Biomolecular condensates located throughout the nucleus and cytoplasm. Created with BioRender.com.
2.2. Multivalent Interactions Promote the Formation of a Phase Separation Network
Recent studies have shown that biomolecular phase separation occurs through multivalency or the capacity to participate in weak multivalent effects that rapidly assemble, disconnect, and recombine. These multiple interactions are facilitated by proteins that embrace multiple folding module domains or intrinsically disordered regions (IDRs) [32] or oligomerization domains [33]. Another type of phase separation protein contains polymeric structural fields, such as DIX domains, which can cross-link to form three-dimensional condensates [34]. These multivalent interactions mainly include two types: one class of intracellular interactions, such as protein–protein, protein–RNA, and RNA–RNA interactions; and another class of weak, instant, multivalent interplay between IDRs, consisting of π–π interactions, cation–anion interactions, dipole–dipole interactions, and π–cation interactions [10][35][36].
In addition, another mechanism, called bridging-induced phase separation (BIPS), has been demonstrated in a number of chromatin-related phase separation phenomena. It has been confirmed that BIPS is the basis for DNA-mediated clustering cohesion [37]. Local bridging along distal segments of DNA molecules is an essential element of BIPS and a characteristic of BIPS that does not exist in other forms of phase separation [38]. In this way, phase separation is induced between chromatin regions (stable or transient) that interact with different types of bridging factors [38].
How do cells use phase separation to respond to changes in cell surroundings? Cells construct and regulate dynamic membraneless organelles through characteristics encoded in intrinsically disordered proteins (IDPs) of related proteins, many of which play central functions in a variety of cell features. IDPs are conformationally flexible, often interacting with their bonding companions via short sequence motifs that reappear within disordered areas, and this multivalent interplay is common in macromolecular complexes [39]. As T-cell receptor components form a cluster of membrane-associated phase separation signals, during T-cell activation, phase separation is driven by the multivalence of LAT, GRB2, SLP-76, Nck, and WASP [40].
3. Mechanisms of Phase Separation in Tumorigenesis
Incorrect or abnormal phase separation of biological macromolecules is closely related to the occurrence of many types of diseases, such as cancer, neurodegenerative diseases, and infectious diseases [41][42][43]. The formation and regulation of aberrant biomolecular condensates is changing the way people think about coping with many diseases, including tumor genesis, diagnosis, and treatment. Researchers are no longer considering highly recurrent point mutations in tumors merely based on the structural visual field but are also considering the condensates involved in these mutations. Generally, combined with current existing research, there are three mechanisms of abnormal biological phase separation leading to disease: (a) The first mechanism is biomacromolecule condensate dysregulation. In cancer, IDR-related signal receptor mutations or chromosome translocations can promote the shape of signal clusters or condensates at transcription or DNA damage repair sites and then change the cell signal cascade, drive abnormal transcription programs or DNA damage repair, and promote cancer cell proliferation [44]. (b) The second mechanism is the changing of the critical catalysts required for phase separation. There is evidence that enzymes can regulate the assembly of biomolecular condensates through posttranslational modification (PTM). For example, DYRK3 is located in condensate and phosphorylates several serine and threonine residue groups in IDRs [45]. During stress recovery, inactivation and activation of DYRK3 are crucial to the formation and dissolution of SG [46]. (c) The third mechanism is the altering of general physicochemical conditions in cells. Cells exposed to stress undergo extreme fluctuations in the levels of ion concentration, osmotic conditions, and pH values, which can change the solubility and interactions of biological macromolecules, resulting in abnormal phase separation [47] (Figure 2).
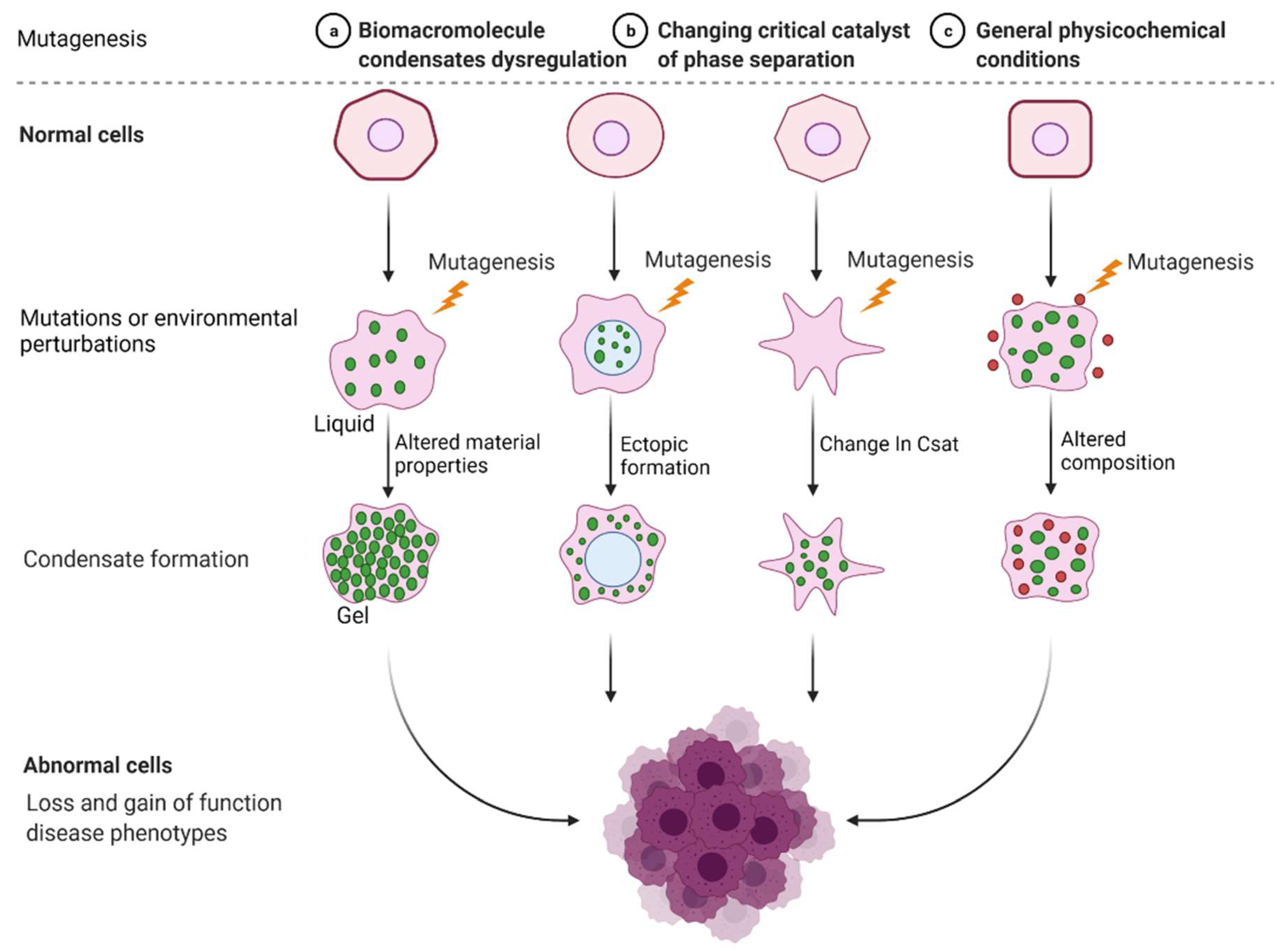
Figure 2. Mechanisms of abnormal phase separation in disease. Theoretical possibilities of how disease phenotypes arise from abnormal phase separation and condensate formation. Created with BioRender.com.
Cancer is the abnormal proliferation of cells in local tissues under the action of various tumorigenic factors in the body. In addition to having unlimited proliferation and multidirectional differentiation potential, many cancer cells have the ability to evade growth repression, engage in replicative immortality, avoid immune destruction, and cause instability of the genome [48], as well as newly discovered tumor features, such as unlocking phenotypic plasticity and reprogramming nonmutational epigenetics [49]. It is worth noting that gene mutations, tumor-promoting inflammation, unlocking phenotypic plasticity, and polymorphic microbiomes increase the possibility of tumors. Gene mutations in cancer often lead to oncogene activity imbalance or inactivation of tumor suppressor genes, thus promoting the carcinogenic process. Despite considerable advances in the understanding of how mutations promote the carcinogenic process, the exact pathogenesis of tumorigenesis remains unclear, as does the mechanism by which tumor cells acquire these features. Phase separation offers a new direction for understanding cancer phenotypes (Figure 3). Generally speaking, it can be divided into two aspects. Dysregulation is driven by phase separation itself. For example, interferon-γ improves tumor sensitivity to immunotherapy by inhibiting YAP phase separation [50]; phase separation of YAP and TAZ participates in activating EMT [51]; and increasing the formation of SGs overcomes stress-induced cancer cell death [52]. In addition to this, carcinogenicity can also be affected by the dysregulation of signaling proteins involved in phase separation. For instance, MYC forms transcription condensates by binding to superenhancers, which lead to VEGF expression and the promotion of angiogenesis [17]. In addition, aberrant phase segregation of the ENL protein can recruit a large number of associated transcription complexes, which lead to genomic rearrangements in cancer [53]. Fusion between promyelocytic leukemia protein (PML) bodies permit telomere lengthening and enable replicative immortality [54], and SPOP mutation inhibits the catabolism of prooncogenic substrates, thus escaping growth inhibition [55]. An increasing number of studies has shown that the process of phase separation cannot be ignored for the progression and treatment of human diseases.
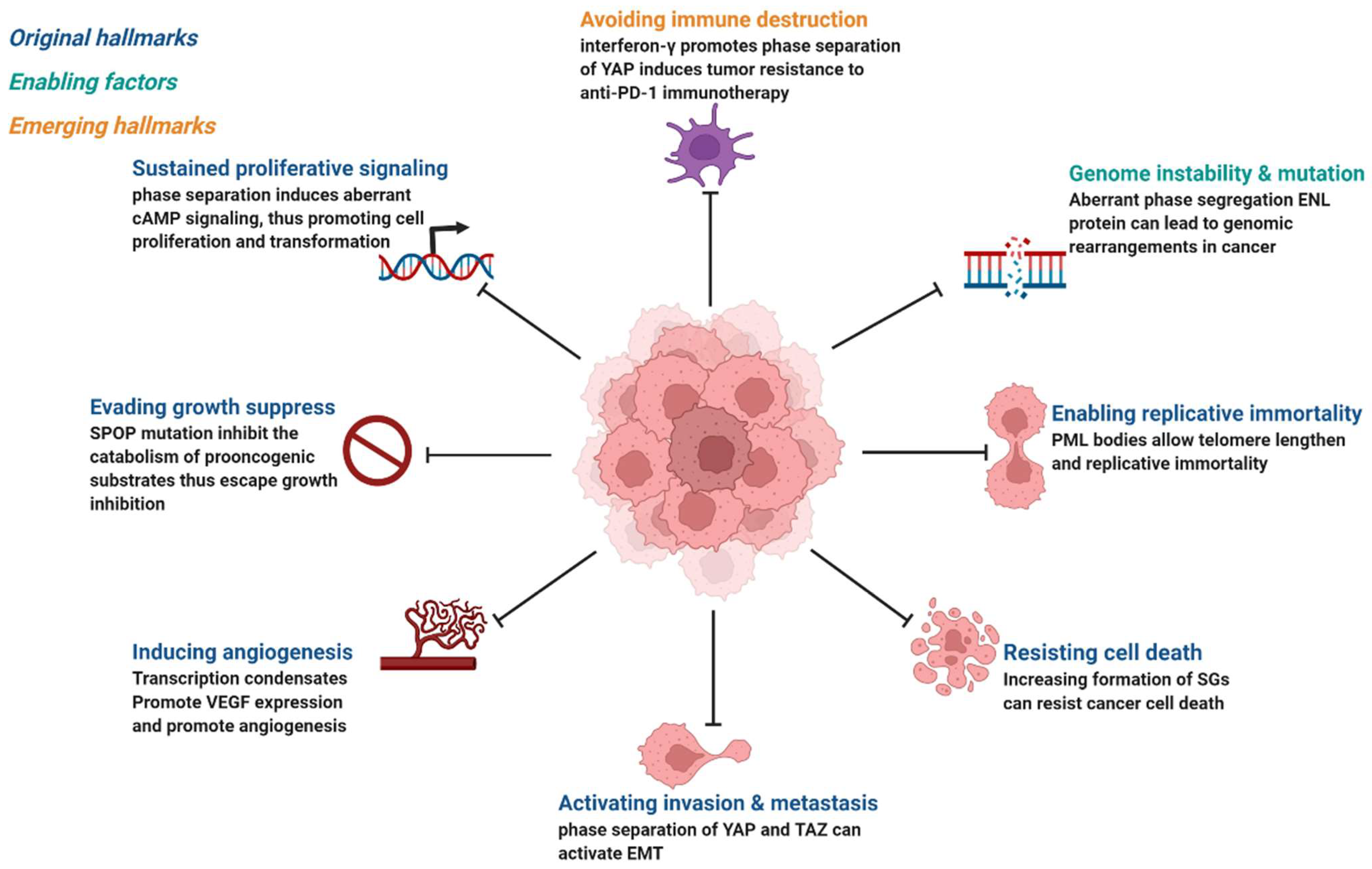
Figure 3. Phase separation abnormalities are involved in most of the processes known as cancer hallmarks. Created with BioRender.com.
Abnormality of phase separation may promote the occurrence of some cancers. For example, one study has directly linked protein phase separation to cancer. In vitro, substrates can trigger phase separation of speckle-type BTB/POZ protein (SPOP) and colocalization in membraneless organelles in cells, and carcinogenic mutations in cancer suppressor SPOP result from interference with phase separation and colocalization in membraneless organelles linked to specific phase separation defects [55]. Cullin3-ring ubiquitin ligase is associated with a variety of solid tumors, and SPOP as its substrate adapter is one of the first cancer-specific related proteins to undergo phase isolation [55][56]. Molecular pathologist Miguel Rivera found a protein associated with Ewing sarcoma. This protein can activate oncogene expression when it accumulates near the genome related to tumorigenesis, and abnormal “phase separation” may promote the aggregation of this protein near these regions, leading to the occurrence of Ewing sarcoma [57]. Moreover, the FUS/EWS/TAF15 (FET) fusion oncoprotein enhances abnormal gene transcription by site-specific phase separation and is an indispensable carcinogenic driver in various human cancers [58][59]. This research reveals that phosphatase protein can undergo phase separation, suggesting that phase separation is a notable means for cells to regulate phosphatase activity. Gene mutation can change the phase separation ability of protein and then change the protein function, leading to the occurrence of human diseases, highlighting the importance of phase separation in human disease occurrence and development [59]. The following is a summary of the various types of cancer condensate formation and the regulation of cancer-associated proteins (Table 1 and Figure 4).
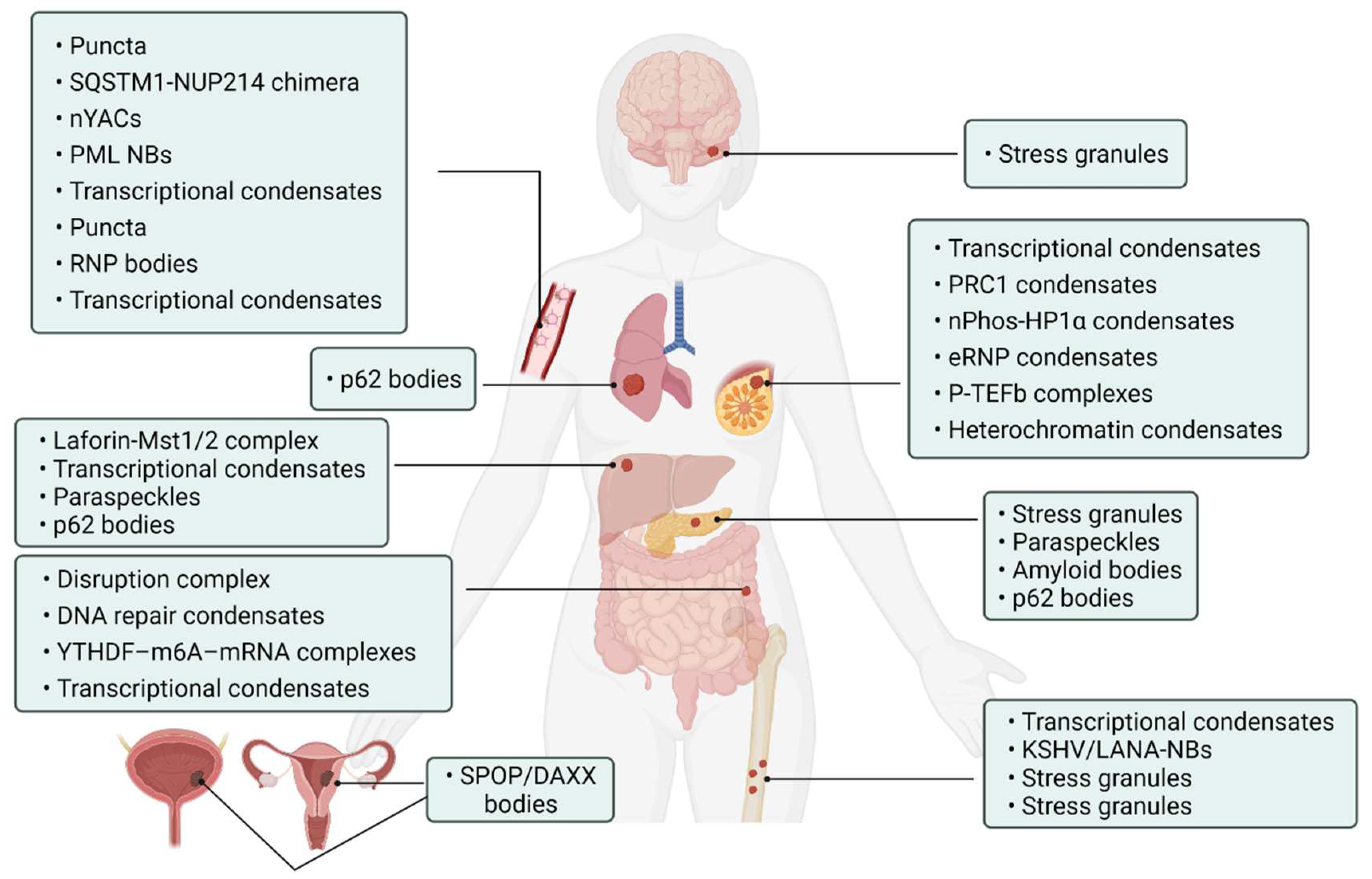
Figure 4. Cancer-related condensates in human cancers. Distribution of various types of cancer and related condensates in the human body. Created with BioRender.com.
Table 1. Condensates in cancer. Abnormal protein phase separations are involved in the progression of various cancers.
| Tumor Types | Proteins | Biomolecular Condensates | Biological Roles | References |
|---|---|---|---|---|
| Hepatocellular Carcinoma | YAP | Laforin-Mst1/2 complex | Block Hippo kinase and accelerate tumorigenesis | [60] |
| YAP, TAZ | Transcriptional condensates | Activate prevalently in cancer | [61][62] | |
| NEAT1_2 | Paraspeckles | Induce transcription of various gene sustained by cancer cells | [63] | |
| p62 | p62 bodies | Induce carcinogenesis | [64] | |
| Lung Cancer | KEAP1/NRF2/p62 | p62 bodies | Increase the risk of tumor genesis | [65] |
| Pancreatic Cancer | KRAS | Stress granules | Improve cancer cell suitability | [52][66] |
| p53 | Paraspeckles | Promote the expression of tumor suppressors | [67] | |
| ACM | Amyloid bodies | Promote tumor tissue growth | [68] | |
| p62 | p62 bodies | Critical in regulating tumorigenesis through autophagy | [69][70][71] | |
| Colorectal Cancer | APC | Disruption complex | Effective β-catenin degradation | [72][73] |
| 53BP1 | DNA repair condensates | Respond to DNA damage | [74] | |
| YTHDF1/2/3 | YTHDF–m6A–mRNA complexes | Weaken mRNA translation | [75][76] | |
| β-catenin | Transcriptional condensates | Wnt factor driving cancer | [77][78] | |
| Leukemia | NUP98 FOs | Puncta | Associated with malignant transformation of hematopoietic cells | [79] |
| NUP214 | SQSTM1-NUP214 chimera | Associated with malignant transformation of hematopoietic cells | [80] | |
| YTHDC1 | nYACs | Maintains mRNA stability and controls cancer cell survival and differentiation | [81] | |
| PML/RARA | PML NBs | Involved in oncogenic signaling | [82][83] | |
| MYB | Transcriptional condensates | Drive oncogenic TAL1 expression | [84] | |
| ENL | Puncta | Regulates oncogenic transcriptional program | [53][85] | |
| NPM1 | RNP bodies | Ribosome biosynthesis | [86][87] | |
| Leukemia/Sarcoma | FUS/TAF15 PLD | Transcriptional condensates | Drive aberrant tumorigenic transcriptional program | [88] |
| Sarcoma | KSHV/LANA | KSHV/LANA-NBs | Cause alterations in gene expression | [89][90] |
| FUS/CHOP | Stress granules | Carcinogenic transformation | [91] | |
| EWS/FLI1 | Transcriptional condensates | Promote gene transcription associated with Ewing’s sarcoma | [92] | |
| YB-1 | Stress granules | Cancer metastatic marker | [93] | |
| Medulloblastoma | DDX3X | Stress granules | Impair global translation | [94][95] |
| Breast Cancer | YAP/TAZ | Transcriptional condensates | Promote expression of target gene | [51][96] |
| CBX2 | PRC1 condensates | Gene suppression | [97][98][99] | |
| HP1α | nPhos-HP1α condensates | Epigenetic regulation | [18][19][100] | |
| ER | eRNP condensates | Synergistic assembly of activated chromosome enhancers | [101] | |
| P-TEFb | P-TEFb complexes | Activate and increase transcription of EMT transcription factors | [102][103] | |
| MeCP2 | Heterochromatin condensates | Chromosome maintenance and transcriptional silence | [104][105] | |
| Prostate/Endometrial cancer | SPOP | SPOP/DAXX bodies | Promote tumor development | [55][56] |
| Other cancers | PARP-1 | DNA damage condensates | Promote DNA damage | [106][107] |
| OCT4 | OCT4-MED1-IDR complex | Control gene transcription | [17][108] | |
| MED1, BRD4 | Transcriptional condensates | Activate gene transcription | [109][110][111] | |
| CDK7 | Transcriptional condensates | Kinase overexpression and targeting in cancer | [112][113] | |
| HSF1 | Transcriptional condensates | Act as “sensors” regulating cell fate | [114][115] | |
| Rad52 | Repair center condensates | DNA repair | [116] | |
| hnRPNA1, FUS, G3NP1/2 | Stress granules | Modulate the stress response | [117][118] | |
| FMRP/CAPRIN1 | FMRP-CAPRIN1 condensates | Control RNA process and translation | [119] |
References
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298.
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498.
- Sachdev, R.; Hondele, M.; Linsenmeier, M.; Vallotton, P.; Mugler, C.F.; Arosio, P.; Weis, K. Pat1 promotes processing body assembly by enhancing the phase separation of the DEAD-box ATPase Dhh1 and RNA. Elife 2019, 8.
- Wheeler, J.R.; Matheny, T.; Jain, S.; Abrisch, R.; Parker, R. Distinct stages in stress granule assembly and disassembly. Elife 2016, 5.
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Van Den Bosch, L.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435.
- Mitrea, D.M.; Kriwacki, R.W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016, 14, 1.
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30.
- Alberti, S.; Dormann, D. Liquid-Liquid Phase Separation in Disease. Annu. Rev. Genet. 2019, 53, 171–194.
- Zhang, H.; Ji, X.; Li, P.; Liu, C.; Lou, J.; Wang, Z.; Wen, W.; Xiao, Y.; Zhang, M.; Zhu, X. Liquid-liquid phase separation in biology: Mechanisms, physiological functions and human diseases. Sci. China Life Sci. 2020, 63, 953–985.
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33.
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434.
- Franzmann, T.M.; Alberti, S. Prion-like low-complexity sequences: Key regulators of protein solubility and phase behavior. J. Biol. Chem. 2019, 294, 7128–7136.
- Su, X.; Ditlev, J.A.; Hui, E.; Xing, W.; Banjade, S.; Okrut, J.; King, D.S.; Taunton, J.; Rosen, M.K.; Vale, R.D. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 2016, 352, 595–599.
- Fujioka, Y.; Alam, J.M.; Noshiro, D.; Mouri, K.; Ando, T.; Okada, Y.; May, A.I.; Knorr, R.L.; Suzuki, K.; Ohsumi, Y.; et al. Phase separation organizes the site of autophagosome formation. Nature 2020, 578, 301–305.
- King, M.R.; Petry, S. Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat. Commun. 2020, 11, 270.
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855.e1816.
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240.
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245.
- Schmidt, H.B.; Görlich, D. Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends Biochem. Sci. 2016, 41, 46–61.
- Xia, S.; Chen, Z.; Shen, C.; Fu, T.M. Higher-order assemblies in immune signaling: Supramolecular complexes and phase separation. Protein Cell 2021, 12, 680–694.
- Yang, P.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345.e328.
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357.
- O’Connell, L.C.; Mowry, K.L. Regulation of spatially restricted gene expression: Linking RNA localization and phase separation. Biochem. Soc. Trans. 2021, 49, 2591–2600.
- Wang, Z.; Zhang, H. Phase Separation, Transition, and Autophagic Degradation of Proteins in Development and Pathogenesis. Trends Cell Biol. 2019, 29, 417–427.
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Jülicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732.
- Buchan, J.R. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014, 11, 1019–1030.
- Lee, C.; Smith, B.A.; Bandyopadhyay, K.; Gjerset, R.A. DNA damage disrupts the p14ARF-B23(nucleophosmin) interaction and triggers a transient subnuclear redistribution of p14ARF. Cancer Res. 2005, 65, 9834–9842.
- Pederson, T. The nucleolus. Cold Spring Harb. Perspect. Biol. 2011, 3.
- Wang, Y.; Chen, L.L. Organization and function of paraspeckles. Essays Biochem. 2020, 64, 875–882.
- Imada, T.; Shimi, T.; Kaiho, A.; Saeki, Y.; Kimura, H. RNA polymerase II condensate formation and association with Cajal and histone locus bodies in living human cells. Genes Cells 2021, 26, 298–312.
- Gomes, E.; Shorter, J. The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294, 7115–7127.
- Dao, T.P.; Kolaitis, R.M.; Kim, H.J.; O’Donovan, K.; Martyniak, B.; Colicino, E.; Hehnly, H.; Taylor, J.P.; Castañeda, C.A. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol. Cell 2018, 69, 965–978.e966.
- Bienz, M. Head-to-Tail Polymerization in the Assembly of Biomolecular Condensates. Cell 2020, 182, 799–811.
- Wang, J.; Choi, J.M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699.e616.
- Li, P.; Banjade, S.; Cheng, H.C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340.
- Ryu, J.K.; Hwang, D.E.; Choi, J.M. Current Understanding of Molecular Phase Separation in Chromosomes. Int. J. Mol. Sci. 2021, 22, 10736.
- Ryu, J.K.; Bouchoux, C.; Liu, H.W.; Kim, E.; Minamino, M.; de Groot, R.; Katan, A.J.; Bonato, A.; Marenduzzo, D.; Michieletto, D.; et al. Bridging-induced phase separation induced by cohesin SMC protein complexes. Sci. Adv. 2021, 7.
- Fung, H.Y.J.; Birol, M.; Rhoades, E. IDPs in macromolecular complexes: The roles of multivalent interactions in diverse assemblies. Curr. Opin. Struct. Biol. 2018, 49, 36–43.
- Ditlev, J.A.; Vega, A.R.; Köster, D.V.; Su, X.; Tani, T.; Lakoduk, A.M.; Vale, R.D.; Mayor, S.; Jaqaman, K.; Rosen, M.K. A composition-dependent molecular clutch between T cell signaling condensates and actin. Elife 2019, 8.
- Gu, X.; Zhuang, A.; Yu, J.; Chai, P.; Jia, R.; Ruan, J. Phase separation drives tumor pathogenesis and evolution: All roads lead to Rome. Oncogene 2022, 41, 1527–1535.
- Loureiro, J.R.; Castro, A.F.; Figueiredo, A.S.; Silveira, I. Molecular Mechanisms in Pentanucleotide Repeat Diseases. Cells 2022, 11, 205.
- Xu, G.; Liu, C.; Zhou, S.; Li, Q.; Feng, Y.; Sun, P.; Feng, H.; Gao, Y.; Zhu, J.; Luo, X.; et al. Viral tegument proteins restrict cGAS-DNA phase separation to mediate immune evasion. Mol. Cell 2021, 81, 2823–2837.e2829.
- Boija, A.; Klein, I.A.; Young, R.A. Biomolecular Condensates and Cancer. Cancer Cell 2021, 39, 174–192.
- Rai, A.K.; Chen, J.X.; Selbach, M.; Pelkmans, L. Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 2018, 559, 211–216.
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013, 152, 791–805.
- Rabouille, C.; Alberti, S. Cell adaptation upon stress: The emerging role of membrane-less compartments. Curr. Opin. Cell Biol. 2017, 47, 34–42.
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674.
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46.
- Yu, M.; Peng, Z.; Qin, M.; Liu, Y.; Wang, J.; Zhang, C.; Lin, J.; Dong, T.; Wang, L.; Li, S.; et al. Interferon-γ induces tumor resistance to anti-PD-1 immunotherapy by promoting YAP phase separation. Mol. Cell 2021, 81, 1216–1230.e1219.
- Lu, Y.; Wu, T.; Gutman, O.; Lu, H.; Zhou, Q.; Henis, Y.I.; Luo, K. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol. 2020, 22, 453–464.
- Grabocka, E.; Bar-Sagi, D. Mutant KRAS Enhances Tumor Cell Fitness by Upregulating Stress Granules. Cell 2016, 167, 1803–1813.e1812.
- Wan, L.; Chong, S.; Xuan, F.; Liang, A.; Cui, X.; Gates, L.; Carroll, T.S.; Li, Y.; Feng, L.; Chen, G.; et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 2020, 577, 121–126.
- Min, J.; Wright, W.E.; Shay, J.W. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 2019, 33, 814–827.
- Bouchard, J.J.; Otero, J.H.; Scott, D.C.; Szulc, E.; Martin, E.W.; Sabri, N.; Granata, D.; Marzahn, M.R.; Lindorff-Larsen, K.; Salvatella, X.; et al. Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol. Cell 2018, 72, 19–36.e18.
- Mani, R.S. The emerging role of speckle-type POZ protein (SPOP) in cancer development. Drug Discov. Today 2014, 19, 1498–1502.
- Boulay, G.; Sandoval, G.J.; Riggi, N.; Iyer, S.; Buisson, R.; Naigles, B.; Awad, M.E.; Rengarajan, S.; Volorio, A.; McBride, M.J.; et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171, 163–178.e119.
- Zuo, L.; Zhang, G.; Massett, M.; Cheng, J.; Guo, Z.; Wang, L.; Gao, Y.; Li, R.; Huang, X.; Li, P.; et al. Loci-specific phase separation of FET fusion oncoproteins promotes gene transcription. Nat. Commun. 2021, 12, 1491.
- Zhu, G.; Xie, J.; Kong, W.; Xie, J.; Li, Y.; Du, L.; Zheng, Q.; Sun, L.; Guan, M.; Li, H.; et al. Phase Separation of Disease-Associated SHP2 Mutants Underlies MAPK Hyperactivation. Cell 2020, 183, 490–502.e418.
- Liu, Q.; Li, J.; Zhang, W.; Xiao, C.; Zhang, S.; Nian, C.; Li, J.; Su, D.; Chen, L.; Zhao, Q.; et al. Glycogen accumulation and phase separation drives liver tumor initiation. Cell 2021, 184, 5559–5576.e5519.
- Cai, D.; Feliciano, D.; Dong, P.; Flores, E.; Gruebele, M.; Porat-Shliom, N.; Sukenik, S.; Liu, Z.; Lippincott-Schwartz, J. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol. 2019, 21, 1578–1589.
- Sengupta, S.; George, R.E. Super-Enhancer-Driven Transcriptional Dependencies in Cancer. Trends Cancer 2017, 3, 269–281.
- Wang, S.; Zhang, Q.; Wang, Q.; Shen, Q.; Chen, X.; Li, Z.; Zhou, Y.; Hou, J.; Xu, B.; Li, N.; et al. NEAT1 paraspeckle promotes human hepatocellular carcinoma progression by strengthening IL-6/STAT3 signaling. Oncoimmunology 2018, 7, e1503913.
- Sun, D.; Wu, R.; Zheng, J.; Li, P.; Yu, L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018, 28, 405–415.
- Cloer, E.W.; Siesser, P.F.; Cousins, E.M.; Goldfarb, D.; Mowrey, D.D.; Harrison, J.S.; Weir, S.J.; Dokholyan, N.V.; Major, M.B. p62-Dependent Phase Separation of Patient-Derived KEAP1 Mutations and NRF2. Mol. Cell Biol. 2018, 38.
- Cox, A.D.; Fesik, S.W.; Kimmelman, A.C.; Luo, J.; Der, C.J. Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 2014, 13, 828–851.
- Mello, S.S.; Sinow, C.; Raj, N.; Mazur, P.K.; Bieging-Rolett, K.; Broz, D.K.; Imam, J.F.C.; Vogel, H.; Wood, L.D.; Sage, J.; et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017, 31, 1095–1108.
- Audas, T.E.; Audas, D.E.; Jacob, M.D.; Ho, J.J.; Khacho, M.; Wang, M.; Perera, J.K.; Gardiner, C.; Bennett, C.A.; Head, T.; et al. Adaptation to Stressors by Systemic Protein Amyloidogenesis. Dev. Cell 2016, 39, 155–168.
- Sánchez-Martín, P.; Saito, T.; Komatsu, M. p62/SQSTM1: ‘Jack of all trades’ in health and cancer. FEBS J. 2019, 286, 8–23.
- Todoric, J.; Antonucci, L.; Di Caro, G.; Li, N.; Wu, X.; Lytle, N.K.; Dhar, D.; Banerjee, S.; Fagman, J.B.; Browne, C.D.; et al. Stress-Activated NRF2-MDM2 Cascade Controls Neoplastic Progression in Pancreas. Cancer Cell 2017, 32, 824–839.e828.
- Ling, J.; Kang, Y.; Zhao, R.; Xia, Q.; Lee, D.F.; Chang, Z.; Li, J.; Peng, B.; Fleming, J.B.; Wang, H.; et al. KrasG12D-induced IKK2/β/NF-κB activation by IL-1α and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell 2012, 21, 105–120.
- Pronobis, M.I.; Rusan, N.M.; Peifer, M. A novel GSK3-regulated APC:Axin interaction regulates Wnt signaling by driving a catalytic cycle of efficient βcatenin destruction. Elife 2015, 4, e08022.
- Li, T.M.; Ren, J.; Husmann, D.; Coan, J.P.; Gozani, O.; Chua, K.F. Multivalent tumor suppressor adenomatous polyposis coli promotes Axin biomolecular condensate formation and efficient β-catenin degradation. Sci. Rep. 2020, 10, 17425.
- Kilic, S.; Lezaja, A.; Gatti, M.; Bianco, E.; Michelena, J.; Imhof, R.; Altmeyer, M. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019, 38, e101379.
- Ries, R.J.; Zaccara, S.; Klein, P.; Olarerin-George, A.; Namkoong, S.; Pickering, B.F.; Patil, D.P.; Kwak, H.; Lee, J.H.; Jaffrey, S.R. m(6)A enhances the phase separation potential of mRNA. Nature 2019, 571, 424–428.
- Gao, Y.; Pei, G.; Li, D.; Li, R.; Shao, Y.; Zhang, Q.C.; Li, P. Multivalent m(6)A motifs promote phase separation of YTHDF proteins. Cell Res. 2019, 29, 767–769.
- Zamudio, A.V.; Dall’Agnese, A.; Henninger, J.E.; Manteiga, J.C.; Afeyan, L.K.; Hannett, N.M.; Coffey, E.L.; Li, C.H.; Oksuz, O.; Sabari, B.R.; et al. Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Mol. Cell 2019, 76, 753–766.e756.
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999.
- Chandra, B.; Michmerhuizen, N.L.; Shirnekhi, H.K.; Tripathi, S.; Pioso, B.J.; Baggett, D.W.; Mitrea, D.M.; Iacobucci, I.; White, M.R.; Chen, J.; et al. Phase Separation Mediates NUP98 Fusion Oncoprotein Leukemic Transformation. Cancer Discov. 2022, 12, 1152–1169.
- Mendes, A.; Fahrenkrog, B. NUP214 in Leukemia: It’s More than Transport. Cells 2019, 8, 76.
- Cheng, Y.; Xie, W.; Pickering, B.F.; Chu, K.L.; Savino, A.M.; Yang, X.; Luo, H.; Nguyen, D.T.; Mo, S.; Barin, E.; et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell 2021, 39, 958–972.e958.
- Dos Santos, G.A.; Kats, L.; Pandolfi, P.P. Synergy against PML-RARa: Targeting transcription, proteolysis, differentiation, and self-renewal in acute promyelocytic leukemia. J. Exp. Med. 2013, 210, 2793–2802.
- Voisset, E.; Moravcsik, E.; Stratford, E.W.; Jaye, A.; Palgrave, C.J.; Hills, R.K.; Salomoni, P.; Kogan, S.C.; Solomon, E.; Grimwade, D. Pml nuclear body disruption cooperates in APL pathogenesis and impairs DNA damage repair pathways in mice. Blood 2018, 131, 636–648.
- Mansour, M.R.; Abraham, B.J.; Anders, L.; Berezovskaya, A.; Gutierrez, A.; Durbin, A.D.; Etchin, J.; Lawton, L.; Sallan, S.E.; Silverman, L.B.; et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 2014, 346, 1373–1377.
- Wan, L.; Wen, H.; Li, Y.; Lyu, J.; Xi, Y.; Hoshii, T.; Joseph, J.K.; Wang, X.; Loh, Y.E.; Erb, M.A.; et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 2017, 543, 265–269.
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697.
- Mitrea, D.M.; Cika, J.A.; Stanley, C.B.; Nourse, A.; Onuchic, P.L.; Banerjee, P.R.; Phillips, A.H.; Park, C.G.; Deniz, A.A.; Kriwacki, R.W. Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun. 2018, 9, 842.
- Tan, A.Y.; Manley, J.L. The TET family of proteins: Functions and roles in disease. J. Mol. Cell Biol. 2009, 1, 82–92.
- Lu, C.; Jain, S.U.; Hoelper, D.; Bechet, D.; Molden, R.C.; Ran, L.; Murphy, D.; Venneti, S.; Hameed, M.; Pawel, B.R.; et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 2016, 352, 844–849.
- Vladimirova, O.; De Leo, A.; Deng, Z.; Wiedmer, A.; Hayden, J.; Lieberman, P.M. Phase separation and DAXX redistribution contribute to LANA nuclear body and KSHV genome dynamics during latency and reactivation. PLoS Pathog. 2021, 17, e1009231.
- Owen, I.; Yee, D.; Wyne, H.; Perdikari, T.M.; Johnson, V.; Smyth, J.; Kortum, R.; Fawzi, N.L.; Shewmaker, F. The oncogenic transcription factor FUS-CHOP can undergo nuclear liquid-liquid phase separation. J. Cell Sci. 2021, 134, jcs258578.
- Chong, S.; Dugast-Darzacq, C.; Liu, Z.; Dong, P.; Dailey, G.M.; Cattoglio, C.; Heckert, A.; Banala, S.; Lavis, L.; Darzacq, X.; et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 2018, 361.
- Somasekharan, S.P.; El-Naggar, A.; Leprivier, G.; Cheng, H.; Hajee, S.; Grunewald, T.G.; Zhang, F.; Ng, T.; Delattre, O.; Evdokimova, V.; et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J. Cell Biol. 2015, 208, 913–929.
- Valentin-Vega, Y.A.; Wang, Y.D.; Parker, M.; Patmore, D.M.; Kanagaraj, A.; Moore, J.; Rusch, M.; Finkelstein, D.; Ellison, D.W.; Gilbertson, R.J.; et al. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci. Rep. 2016, 6, 25996.
- Hondele, M.; Sachdev, R.; Heinrich, S.; Wang, J.; Vallotton, P.; Fontoura, B.M.A.; Weis, K. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 2019, 573, 144–148.
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP and TAZ: A signalling hub of the tumour microenvironment. Nat. Rev. Cancer 2019, 19, 454–464.
- Plys, A.J.; Davis, C.P.; Kim, J.; Rizki, G.; Keenen, M.M.; Marr, S.K.; Kingston, R.E. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019, 33, 799–813.
- Tatavosian, R.; Kent, S.; Brown, K.; Yao, T.; Duc, H.N.; Huynh, T.N.; Zhen, C.Y.; Ma, B.; Wang, H.; Ren, X. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019, 294, 1451–1463.
- Clermont, P.L.; Sun, L.; Crea, F.; Thu, K.L.; Zhang, A.; Parolia, A.; Lam, W.L.; Helgason, C.D. Genotranscriptomic meta-analysis of the Polycomb gene CBX2 in human cancers: Initial evidence of an oncogenic role. Br. J. Cancer 2014, 111, 1663–1672.
- Lieberthal, J.G.; Kaminsky, M.; Parkhurst, C.N.; Tanese, N. The role of YY1 in reduced HP1alpha gene expression in invasive human breast cancer cells. Breast Cancer Res. 2009, 11, R42.
- Nair, S.J.; Yang, L.; Meluzzi, D.; Oh, S.; Yang, F.; Friedman, M.J.; Wang, S.; Suter, T.; Alshareedah, I.; Gamliel, A.; et al. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol. 2019, 26, 193–203.
- Ji, X.; Lu, H.; Zhou, Q.; Luo, K. LARP7 suppresses P-TEFb activity to inhibit breast cancer progression and metastasis. Elife 2014, 3, e02907.
- Lu, H.; Yu, D.; Hansen, A.S.; Ganguly, S.; Liu, R.; Heckert, A.; Darzacq, X.; Zhou, Q. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018, 558, 318–323.
- Li, C.H.; Coffey, E.L.; Dall’Agnese, A.; Hannett, N.M.; Tang, X.; Henninger, J.E.; Platt, J.M.; Oksuz, O.; Zamudio, A.V.; Afeyan, L.K.; et al. MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature 2020, 586, 440–444.
- Neupane, M.; Clark, A.P.; Landini, S.; Birkbak, N.J.; Eklund, A.C.; Lim, E.; Culhane, A.C.; Barry, W.T.; Schumacher, S.E.; Beroukhim, R.; et al. MECP2 Is a Frequently Amplified Oncogene with a Novel Epigenetic Mechanism That Mimics the Role of Activated RAS in Malignancy. Cancer Discov. 2016, 6, 45–58.
- Singatulina, A.S.; Hamon, L.; Sukhanova, M.V.; Desforges, B.; Joshi, V.; Bouhss, A.; Lavrik, O.I.; Pastré, D. PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell Rep. 2019, 27, 1809–1821.e1805.
- Altmeyer, M.; Neelsen, K.J.; Teloni, F.; Pozdnyakova, I.; Pellegrino, S.; Grøfte, M.; Rask, M.D.; Streicher, W.; Jungmichel, S.; Nielsen, M.L.; et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 2015, 6, 8088.
- Kumar, S.M.; Liu, S.; Lu, H.; Zhang, H.; Zhang, P.J.; Gimotty, P.A.; Guerra, M.; Guo, W.; Xu, X. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene 2012, 31, 4898–4911.
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361.
- Nagalingam, A.; Tighiouart, M.; Ryden, L.; Joseph, L.; Landberg, G.; Saxena, N.K.; Sharma, D. Med1 plays a critical role in the development of tamoxifen resistance. Carcinogenesis 2012, 33, 918–930.
- Russo, J.W.; Nouri, M.; Balk, S.P. Androgen Receptor Interaction with Mediator Complex Is Enhanced in Castration-Resistant Prostate Cancer by CDK7 Phosphorylation of MED1. Cancer Discov. 2019, 9, 1490–1492.
- Klein, I.A.; Boija, A.; Afeyan, L.K.; Hawken, S.W.; Fan, M.; Dall’Agnese, A.; Oksuz, O.; Henninger, J.E.; Shrinivas, K.; Sabari, B.R.; et al. Partitioning of cancer therapeutics in nuclear condensates. Science 2020, 368, 1386–1392.
- Kwiatkowski, N.; Zhang, T.; Rahl, P.B.; Abraham, B.J.; Reddy, J.; Ficarro, S.B.; Dastur, A.; Amzallag, A.; Ramaswamy, S.; Tesar, B.; et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014, 511, 616–620.
- Gaglia, G.; Rashid, R.; Yapp, C.; Joshi, G.N.; Li, C.G.; Lindquist, S.L.; Sarosiek, K.A.; Whitesell, L.; Sorger, P.K.; Santagata, S. HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat. Cell Biol. 2020, 22, 151–158.
- Carpenter, R.L.; Gökmen-Polar, Y. HSF1 as a Cancer Biomarker and Therapeutic Target. Curr. Cancer Drug Targets 2019, 19, 515–524.
- Oshidari, R.; Huang, R.; Medghalchi, M.; Tse, E.Y.W.; Ashgriz, N.; Lee, H.O.; Wyatt, H.; Mekhail, K. DNA repair by Rad52 liquid droplets. Nat. Commun. 2020, 11, 695.
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679.
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163, 123–133.
- Kim, T.H.; Tsang, B.; Vernon, R.M.; Sonenberg, N.; Kay, L.E.; Forman-Kay, J.D. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 2019, 365, 825–829.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.5K
Revisions:
2 times
(View History)
Update Date:
06 Dec 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





