
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Dafni Moriki | -- | 3275 | 2022-11-09 19:08:08 | | | |
| 2 | Peter Tang | Meta information modification | 3275 | 2022-11-10 02:20:41 | | |
Video Upload Options
Cow’s milk allergy (CMA) is the most prevalent food allergy (FA) in infancy and early childhood and can be present with various clinical phenotypes. The significant increase in FA rates recorded has been associated with environmental and lifestyle changes that limit microbial exposure in early life and induce changes in gut microbiome composition. Gut microbiome is a diverse community of microbes that colonize the gastrointestinal tract (GIT) and perform beneficial functions for the host. This complex ecosystem interacts with the immune system and has a pivotal role in the development of oral tolerance to food antigens. Emerging evidence indicates that alterations of the gut microbiome (dysbiosis) in early life cause immune dysregulation and render the host susceptible to immune-mediated diseases later in life.
1. Introduction
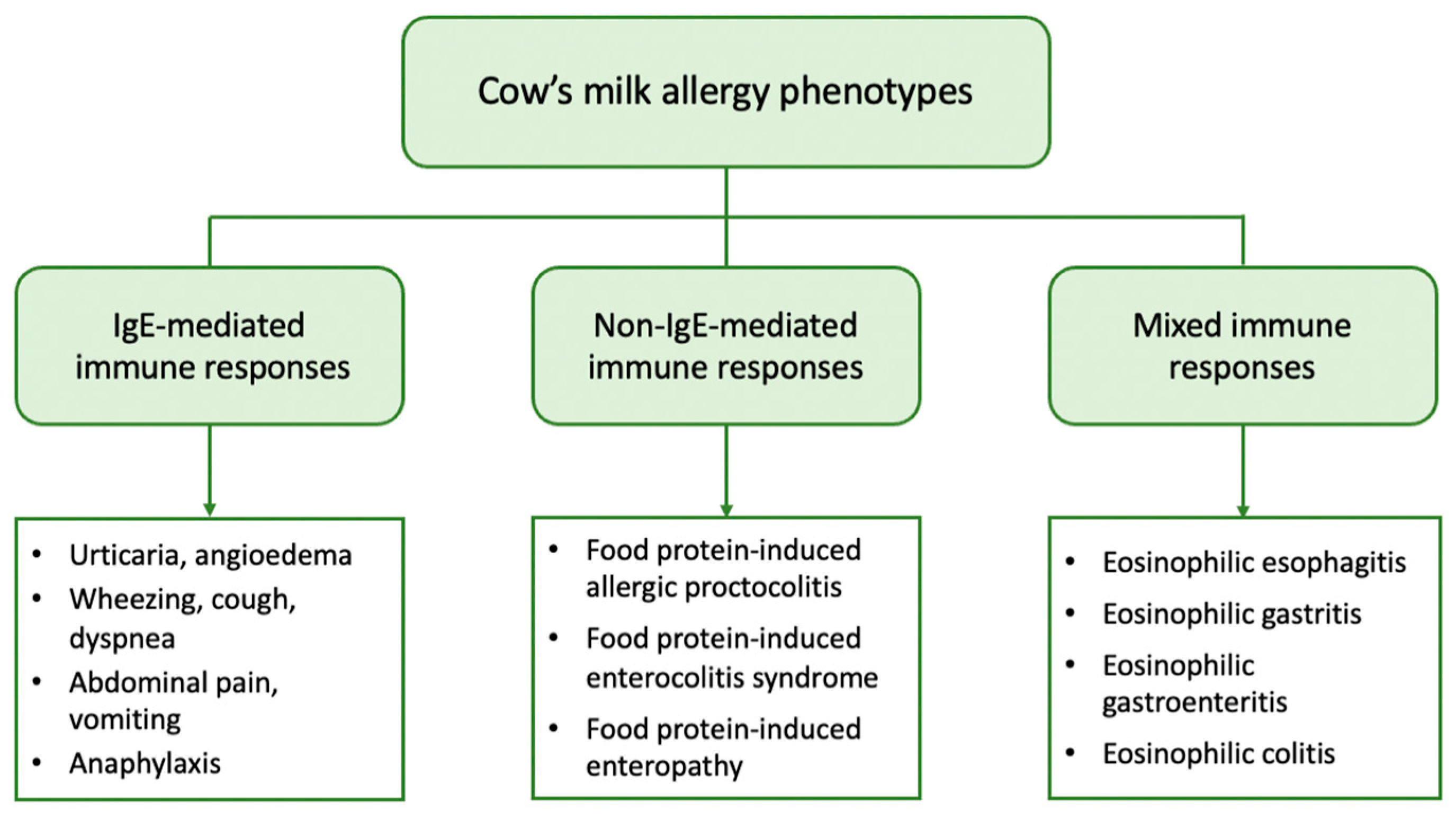
2. Clinical Phenotypes of CMA
3. Gut Microbiome and the Factors Influencing Its Development in Early Life
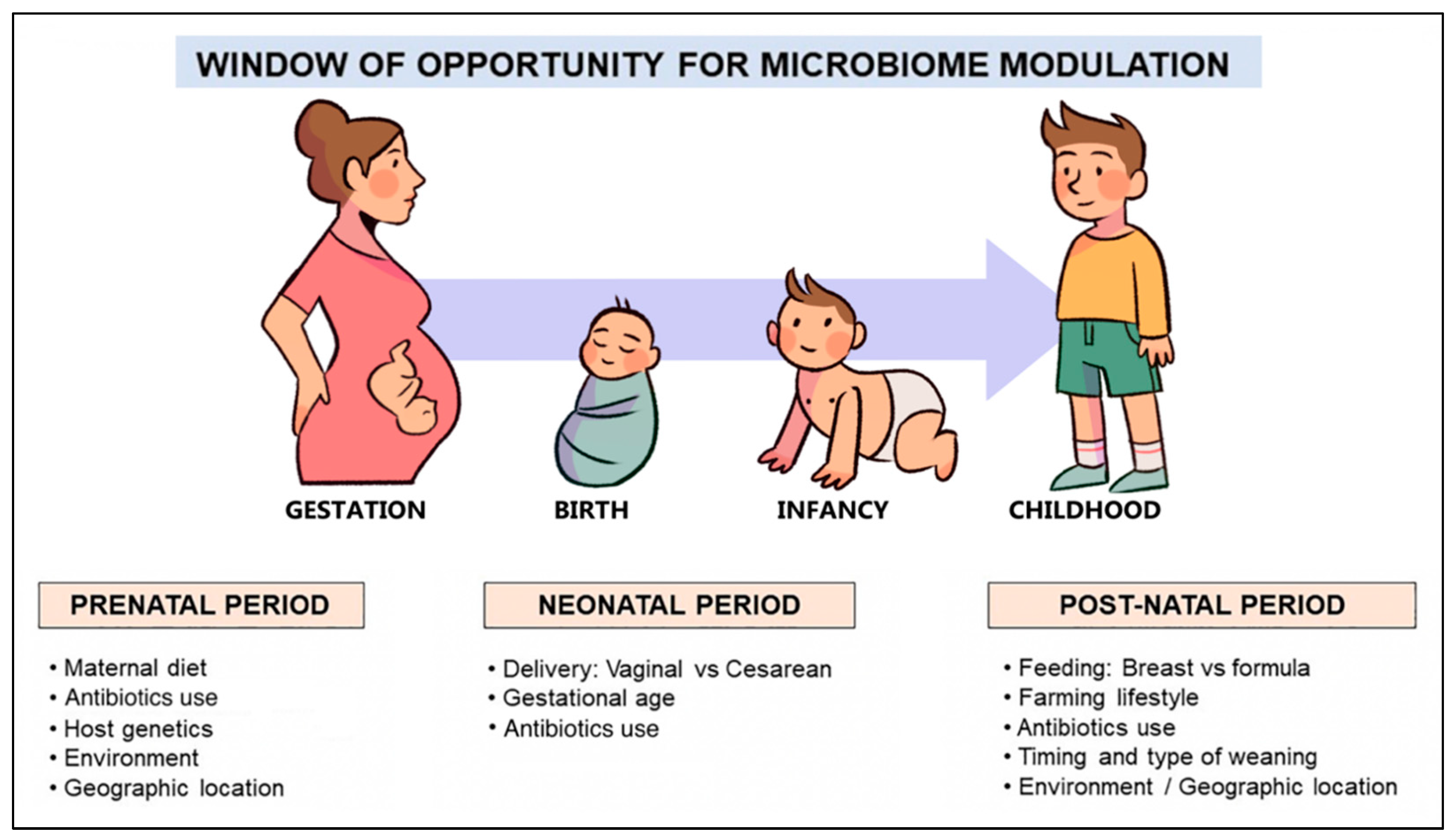
4. The Role of the Gut Microbiome in the Development of FA
References
- Sicherer, S.H.; Sampson, H.A. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J. Allergy Clin. Immunol. 2014, 133, 291–307.
- Savage, J.; Johns, C.B. Food allergy: Epidemiology and natural history. Immunol. Allergy Clin. N. Am. 2015, 35, 45–59.
- Lifschitz, C.; Szajewska, H. Cow’s milk allergy: Evidence-based diagnosis and management for the practitioner. Eur. J. Pediatr. 2015, 174, 141–150.
- Nwaru, B.I.; Hickstein, L.; Panesar, S.S.; Roberts, G.; Muraro, A.; Sheikh, A. Prevalence of common food allergies in Europe: A systematic review and meta-analysis. Allergy 2014, 69, 992–1007.
- Schoemaker, A.A.; Sprikkelman, A.B.; Grimshaw, K.E.; Roberts, G.; Grabenhenrich, L.; Rosenfeld, L.; Siegert, S.; Dubakiene, R.; Rudzeviciene, O.; Reche, M.; et al. Incidence and natural history of challenge-proven cow’s milk allergy in European children--EuroPrevall birth cohort. Allergy 2015, 70, 963–972.
- Rona, R.J.; Keil, T.; Summers, C.; Gislason, D.; Zuidmeer, L.; Sodergren, E.; Sigurdardottir, S.T.; Lindner, T.; Goldhahn, K.; Dahlstrom, J.; et al. The prevalence of food allergy: A meta-analysis. J. Allergy Clin. Immunol. 2007, 120, 638–646.
- Katz, Y.; Rajuan, N.; Goldberg, M.R.; Eisenberg, E.; Heyman, E.; Cohen, A.; Leshno, M. Early exposure to cow’s milk protein is protective against IgE-mediated cow’s milk protein allergy. J. Allergy Clin. Immunol. 2010, 126, 77–82.
- García-Ara, M.C.; Boyano-Martínez, M.T.; Díaz-Pena, J.M.; Martín-Muñoz, M.F.; Martín-Esteban, M. Cow’s milk-specific immunoglobulin E levels as predictors of clinical reactivity in the follow-up of the cow’s milk allergy infants. Clin. Exp. Allergy 2004, 34, 866–870.
- Fiocchi, A.; Terracciano, L.; Bouygue, G.R.; Veglia, F.; Sarratud, T.; Martelli, A.; Restani, P. Incremental prognostic factors associated with cow’s milk allergy outcomes in infant and child referrals: The Milan Cow’s Milk Allergy Cohort study. Ann. Allergy Asthma Immunol. 2008, 101, 166–173.
- Skripak, J.M.; Matsui, E.C.; Mudd, K.; Wood, R.A. The natural history of IgE-mediated cow’s milk allergy. J. Allergy Clin. Immunol. 2007, 120, 1172–1177.
- Tsabouri, S.; Douros, K.; Priftis, K.N. Cow’s milk allergenicity. Endocr. Metab. Immune Disord. Drug Targets 2014, 14, 16–26.
- Yeung, J.P.; Kloda, L.A.; McDevitt, J.; Ben-Shoshan, M.; Alizadehfar, R. Oral immunotherapy for milk allergy. Cochrane Database Syst. Rev. 2012, 11, CD009542.
- Vandenplas, Y. Prevention and Management of Cow’s Milk Allergy in Non-Exclusively Breastfed Infants. Nutrients 2017, 9, 731.
- McBurney, M.I.; Davis, C.; Fraser, C.M.; Schneeman, B.O.; Huttenhower, C.; Verbeke, K.; Walter, J.; Latulippe, M.E. Establishing What Constitutes a Healthy Human Gut Microbiome: State of the Science, Regulatory Considerations, and Future Directions. J. Nutr. 2019, 149, 1882–1895.
- Moran, N.A.; Ochman, H.; Hammer, T.J. Evolutionary and ecological consequences of gut microbial communities. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 451–475.
- Young, V.B. The role of the microbiome in human health and disease: An introduction for clinicians. BMJ Br. Med. J. 2017, 356, j831.
- Fiocchi, A.; Schünemann, H.J.; Brozek, J.; Restani, P.; Beyer, K.; Troncone, R.; Martelli, A.; Terracciano, L.; Bahna, S.L.; Rancé, F.; et al. Diagnosis and Rationale for Action Against Cow’s Milk Allergy (DRACMA): A summary report. J. Allergy Clin. Immunol. 2010, 126, 1119–1128.
- Sampson, H.A. Food allergy. Part 1: Immunopathogenesis and clinical disorders. J. Allergy Clin. Immunol. 1999, 103, 717–728.
- Vickery, B.P.; Chin, S.; Burks, A.W. Pathophysiology of food allergy. Pediatr. Clin. N. Am. 2011, 58, 363–376.
- Luyt, D.; Ball, H.; Makwana, N.; Green, M.R.; Bravin, K.; Nasser, S.M.; Clark, A.T. BSACI guideline for the diagnosis and management of cow’s milk allergy. Clin. Exp. Allergy 2014, 44, 642–672.
- Cianferoni, A. Non-IgE Mediated Food Allergy. Curr. Pediatr. Rev. 2020, 16, 95–105.
- Nowak-Węgrzyn, A.; Katz, Y.; Mehr, S.S.; Koletzko, S. Non-IgE-mediated gastrointestinal food allergy. J. Allergy Clin. Immunol. 2015, 135, 1114–1124.
- Lin, X.P.; Magnusson, J.; Ahlstedt, S.; Dahlman-Höglund, A.; Hanson, L.L.; Magnusson, O.; Bengtsson, U.; Telemo, E. Local allergic reaction in food-hypersensitive adults despite a lack of systemic food-specific IgE. J. Allergy Clin. Immunol. 2002, 109, 879–887.
- Katz, Y.; Goldberg, M.R.; Rajuan, N.; Cohen, A.; Leshno, M. The prevalence and natural course of food protein-induced enterocolitis syndrome to cow’s milk: A large-scale, prospective population-based study. J. Allergy Clin. Immunol. 2011, 127, 647–653.
- Alonso, S.B.; Ezquiaga, J.G.; Berzal, P.T.; Tardón, S.D.; San José, M.M.; López, P.A.; Bermejo, T.B.; Teruel, S.Q.; Echeverría Zudaire, L. Food protein-induced enterocolitis syndrome: Increased prevalence of this great unknown-results of the PREVALE study. J. Allergy Clin. Immunol. 2019, 143, 430–433.
- Leonard, S.A.; Pecora, V.; Fiocchi, A.G.; Nowak-Wegrzyn, A. Food protein-induced enterocolitis syndrome: A review of the new guidelines. World Allergy Organ. J. 2018, 11, 4.
- Labrosse, R.; Graham, F.; Caubet, J.C. Non-IgE-Mediated Gastrointestinal Food Allergies in Children: An Update. Nutrients 2020, 12, 2086.
- Gonsalves, N. Eosinophilic Gastrointestinal Disorders. Clin. Rev. Allergy Immunol. 2019, 57, 272–285.
- Nutten, S. Atopic dermatitis: Global epidemiology and risk factors. Ann. Nutr. Metab. 2015, 66 (Suppl. 1), 8–16.
- Zhu, T.H.; Zhu, T.R.; Tran, K.A.; Sivamani, R.K.; Shi, V.Y. Epithelial barrier dysfunctions in atopic dermatitis: A skin-gut-lung model linking microbiome alteration and immune dysregulation. Br. J. Dermatol. 2018, 179, 570–581.
- Bergmann, M.M.; Caubet, J.C.; Boguniewicz, M.; Eigenmann, P.A. Evaluation of food allergy in patients with atopic dermatitis. J. Allergy Clin. Immunol. Pract. 2013, 1, 22–28.
- Marzuillo, P.; Guarino, S.; Perrone, L. Atopic eczema could be a cause and not an effect of cow’s milk protein allergy. J. Pediatr. Gastroenterol. Nutr. 2014, 58, e23.
- Misselwitz, B.; Butter, M.; Verbeke, K.; Fox, M.R. Update on lactose malabsorption and intolerance: Pathogenesis, diagnosis and clinical management. Gut 2019, 68, 2080–2091.
- Fassio, F.; Facioni, M.S.; Guagnini, F. Lactose Maldigestion, Malabsorption, and Intolerance: A Comprehensive Review with a Focus on Current Management and Future Perspectives. Nutrients 2018, 10, 1599.
- Storhaug, C.L.; Fosse, S.K.; Fadnes, L.T. Country, regional, and global estimates for lactose malabsorption in adults: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2017, 2, 738–746.
- Heine, R.G.; AlRefaee, F.; Bachina, P.; De Leon, J.C.; Geng, L.; Gong, S.; Madrazo, J.A.; Ngamphaiboon, J.; Ong, C.; Rogacion, J.M. Lactose intolerance and gastrointestinal cow’s milk allergy in infants and children—Common misconceptions revisited. World Allergy Organ. J. 2017, 10, 41.
- Catanzaro, R.; Sciuto, M.; Marotta, F. Lactose intolerance: An update on its pathogenesis, diagnosis, and treatment. Nutr. Res. 2021, 89, 23–34.
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638.
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180.
- Adak, A.; Khan, M.R. An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. 2019, 76, 473–493.
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703.
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227.
- Gosalbes, M.J.; Llop, S.; Vallès, Y.; Moya, A.; Ballester, F.; Francino, M.P. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 2013, 43, 198–211.
- Moles, L.; Gómez, M.; Heilig, H.; Bustos, G.; Fuentes, S.; de Vos, W.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS ONE 2013, 8, e66986.
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra65.
- Robertson, R.C.; Manges, A.R.; Finlay, B.B.; Prendergast, A.J. The Human Microbiome and Child Growth—First 1000 Days and Beyond. Trends Microbiol. 2019, 27, 131–147.
- Francino, M.P. Early development of the gut microbiota and immune health. Pathogens 2014, 3, 769–790.
- Francino, M.P. Birth Mode-Related Differences in Gut Microbiota Colonization and Immune System Development. Ann. Nutr. Metab. 2018, 73 (Suppl. 3), 12–16.
- Peroni, D.G.; Nuzzi, G.; Trambusti, I.; Di Cicco, M.E.; Comberiati, P. Microbiome Composition and Its Impact on the Development of Allergic Diseases. Front. Immunol. 2020, 11, 700.
- Tsabouri, S.; Priftis, K.N.; Chaliasos, N.; Siamopoulou, A. Modulation of gut microbiota downregulates the development of food allergy in infancy. Allergol. Immunopathol. 2014, 42, 69–77.
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975.
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019, 574, 117–121.
- Kristensen, K.; Henriksen, L. Cesarean section and disease associated with immune function. J. Allergy Clin. Immunol. 2016, 137, 587–590.
- Kolokotroni, O.; Middleton, N.; Gavatha, M.; Lamnisos, D.; Priftis, K.N.; Yiallouros, P.K. Asthma and atopy in children born by caesarean section: Effect modification by family history of allergies—A population based cross-sectional study. BMC Pediatr. 2012, 12, 179.
- Karlström, A.; Lindgren, H.; Hildingsson, I. Maternal and infant outcome after caesarean section without recorded medical indication: Findings from a Swedish case-control study. BJOG Int. J. Obstet. Gynaecol. 2013, 120, 479–486.
- Magne, F.; Abély, M.; Boyer, F.; Morville, P.; Pochart, P.; Suau, A. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol. Ecol. 2006, 57, 128–138.
- Arboleya, S.; Solís, G.; Fernández, N.; de los Reyes-Gavilán, C.G.; Gueimonde, M. Facultative to strict anaerobes ratio in the preterm infant microbiota: A target for intervention? Gut Microbes 2012, 3, 583–588.
- Jacquot, A.; Neveu, D.; Aujoulat, F.; Mercier, G.; Marchandin, H.; Jumas-Bilak, E.; Picaud, J.C. Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients. J. Pediatr. 2011, 158, 390–396.
- Mai, V.; Torrazza, R.M.; Ukhanova, M.; Wang, X.; Sun, Y.; Li, N.; Shuster, J.; Sharma, R.; Hudak, M.L.; Neu, J. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS ONE 2013, 8, e52876.
- Cuna, A.; Morowitz, M.J.; Ahmed, I.; Umar, S.; Sampath, V. Dynamics of the preterm gut microbiome in health and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G411–G419.
- Lyons, K.E.; Ryan, C.A.; Dempsey, E.M.; Ross, R.P.; Stanton, C. Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients 2020, 12, 1039.
- Levin, A.M.; Sitarik, A.R.; Havstad, S.L.; Fujimura, K.E.; Wegienka, G.; Cassidy-Bushrow, A.E.; Kim, H.; Zoratti, E.M.; Lukacs, N.W.; Boushey, H.A.; et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci. Rep. 2016, 6, 31775.
- Francino, M.P. Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Front. Microbiol. 2015, 6, 1543.
- Tanaka, S.; Kobayashi, T.; Songjinda, P.; Tateyama, A.; Tsubouchi, M.; Kiyohara, C.; Shirakawa, T.; Sonomoto, K.; Nakayama, J. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol. Med. Microbiol. 2009, 56, 80–87.
- Fouhy, F.; Guinane, C.M.; Hussey, S.; Wall, R.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C.; et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob. Agents Chemother. 2012, 56, 5811–5820.
- Laforest-Lapointe, I.; Arrieta, M.C. Patterns of Early-Life Gut Microbial Colonization during Human Immune Development: An Ecological Perspective. Front. Immunol. 2017, 8, 788.
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; van den Brandt, P.A.; Stobberingh, E.E. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006, 118, 511–521.
- Iweala, O.I.; Nagler, C.R. Immune privilege in the gut: The establishment and maintenance of non-responsiveness to dietary antigens and commensal flora. Immunol. Rev. 2006, 213, 82–100.
- Rodriguez, B.; Prioult, G.; Bibiloni, R.; Nicolis, I.; Mercenier, A.; Butel, M.J.; Waligora-Dupriet, A.J. Germ-free status and altered caecal subdominant microbiota are associated with a high susceptibility to cow’s milk allergy in mice. FEMS Microbiol. Ecol. 2011, 76, 133–144.
- Morin, S.; Bernard, H.; Przybylski-Nicaise, L.; Corthier, G.; Rabot, S.; Wal, J.M.; Hazebrouck, S. Allergenic and immunogenic potential of cow’s milk β-lactoglobulin and caseins evidenced without adjuvant in germ-free mice. Mol. Nutr. Food Res. 2011, 55, 1700–1707.
- Feehley, T.; Plunkett, C.H.; Bao, R.; Choi Hong, S.M.; Culleen, E.; Belda-Ferre, P.; Campbell, E.; Aitoro, R.; Nocerino, R.; Paparo, L.; et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 2019, 25, 448–453.
- Satitsuksanoa, P.; Jansen, K.; Głobińska, A.; van de Veen, W.; Akdis, M. Regulatory Immune Mechanisms in Tolerance to Food Allergy. Front. Immunol. 2018, 9, 2939.
- Lyons, A.; O’Mahony, D.; O’Brien, F.; MacSharry, J.; Sheil, B.; Ceddia, M.; Russell, W.M.; Forsythe, P.; Bienenstock, J.; Kiely, B.; et al. Bacterial strain-specific induction of Foxp3+ T regulatory cells is protective in murine allergy models. Clin. Exp. Allergy 2010, 40, 811–819.
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341.
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450.
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573.
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455.
- Cao, S.; Feehley, T.J.; Nagler, C.R. The role of commensal bacteria in the regulation of sensitization to food allergens. FEBS Lett. 2014, 588, 4258–4266.
- Stefka, A.T.; Feehley, T.; Tripathi, P.; Qiu, J.; McCoy, K.; Mazmanian, S.K.; Tjota, M.Y.; Seo, G.Y.; Cao, S.; Theriault, B.R.; et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA 2014, 111, 13145–13150.
- Smeekens, J.M.; Johnson-Weaver, B.T.; Hinton, A.L.; Azcarate-Peril, M.A.; Moran, T.P.; Immormino, R.M.; Kesselring, J.R.; Steinbach, E.C.; Orgel, K.A.; Staats, H.F.; et al. Fecal IgA, Antigen Absorption, and Gut Microbiome Composition Are Associated With Food Antigen Sensitization in Genetically Susceptible Mice. Front. Immunol. 2020, 11, 599637.
- Corthésy, B. Multi-faceted functions of secretory IgA at mucosal surfaces. Front. Immunol. 2013, 4, 185.
- Zhang, B.; Liu, E.; Gertie, J.A.; Joseph, J.; Xu, L.; Pinker, E.Y.; Waizman, D.A.; Catanzaro, J.; Hamza, K.H.; Lahl, K.; et al. Divergent T follicular helper cell requirement for IgA and IgE production to peanut during allergic sensitization. Sci. Immunol. 2020, 5, eaay2754.
- León, E.D.; Francino, M.P. Roles of Secretory Immunoglobulin A in Host-Microbiota Interactions in the Gut Ecosystem. Front. Microbiol. 2022, 13, 880484.




