
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Donatella Farini | -- | 3960 | 2022-10-28 15:13:07 | | | |
| 2 | Lindsay Dong | -29 word(s) | 3931 | 2022-10-30 16:27:04 | | |
Video Upload Options
Meiosis is the unique division of germ cells resulting in the recombination of the maternal and paternal genomes and the production of haploid gametes. In mammals, it begins during the fetal life in females and during puberty in males. In both cases, entering meiosis requires a timely switch from the mitotic to the meiotic cell cycle and the transition from a potential pluripotent status to meiotic differentiation. Revealing the molecular mechanisms underlying these interrelated processes represents the essence in understanding the beginning of meiosis. Meiosis facilitates diversity across individuals and acts as a fundamental driver of evolution. Major differences between sexes and among species complicate the understanding of how meiosis begins.
1. Introduction
2. Molecules and Conditions for Meiotic Beginning
2.1. Intrinsic Factors
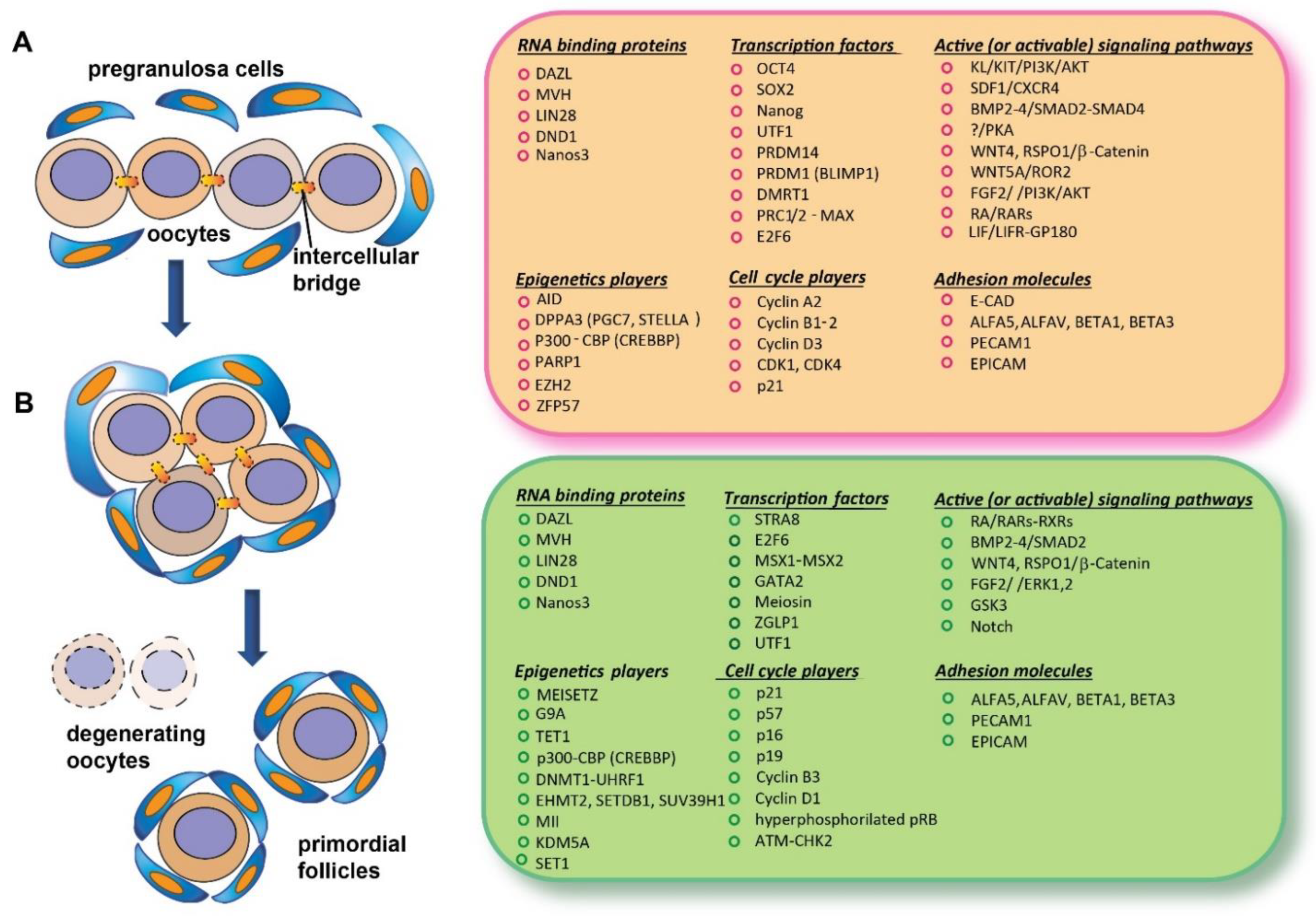
2.1.1. Genome-Wide Reprogramming in Premeiotic PGCs
2.1.2. DAZL and MVH
2.2. Extrinsic Factors
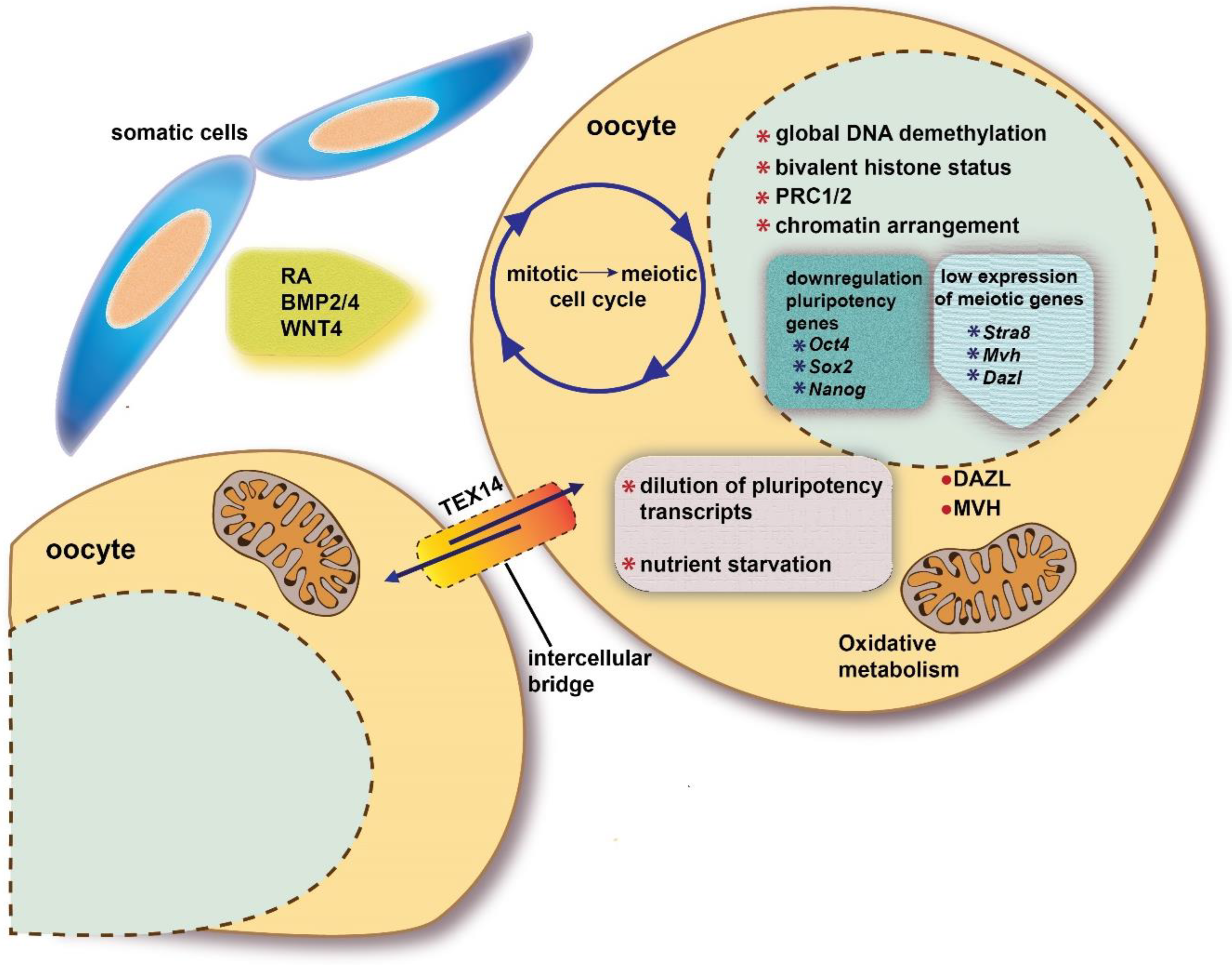
2.2.1. The Switch from Mitotic to Meiotic Cell Cycle
2.2.2. Retinoic Acid
2.2.3. STRA8
2.3. Other Factors
2.3.1. Inhibitory and Cooperative Factors
2.3.2. Nutrient and Metabolic Factors
References
- Hajkova, P.; Ancelin, K.; Waldmann, T.; Lacoste, N.; Lange, U.C.; Cesari, F.; Lee, C.; Almouzni, G.; Schneider, R.; Surani, M.A. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 2008, 452, 877–881.
- Kafri, T.; Ariel, M.; Brandeis, M.; Shemer, R.; Urven, L.; McCarrey, J.; Cedar, H.; Razin, A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992, 6, 705–714.
- Leitch, H.G.; Tang, W.W.; Surani, M.A. Primordial germ-cell development and epigenetic reprogramming in mammals. Curr. Top. Dev. Biol. 2013, 104, 149–187.
- Seisenberger, S.; Andrews, S.; Krueger, F.; Arand, J.; Walter, J.; Santos, F.; Popp, C.; Thienpont, B.; Dean, W.; Reik, W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 2012, 48, 849–862.
- Maatouk, D.M.; Kellam, L.D.; Mann, M.R.; Lei, H.; Li, E.; Bartolomei, M.S.; Resnick, J.L. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development 2006, 133, 3411–3418.
- Popp, C.; Dean, W.; Feng, S.; Cokus, S.J.; Andrews, S.; Pellegrini, M.; Jacobsen, S.E.; Reik, W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 2010, 463, 1101–1105.
- Ciccarone, F.; Klinger, F.G.; Catizone, A.; Calabrese, R.; Zampieri, M.; Bacalini, M.G.; De Felici, M.; Caiafa, P. Poly(ADP-ribosyl)ation acts in the DNA demethylation of mouse primordial germ cells also with DNA damage-independent roles. PLoS ONE 2012, 7, e46927.
- Hajkova, P.; Jeffries, S.J.; Lee, C.; Miller, N.; Jackson, S.P.; Surani, M.A. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 2010, 329, 78–82.
- Kagiwada, S.; Kurimoto, K.; Hirota, T.; Yamaji, M.; Saitou, M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 2013, 32, 340–353.
- Tang, W.W.C.; Dietmann, S.; Irie, N.; Leitch, H.G.; Floros, V.I.; Bradshaw, C.R.; Hackett, J.A.; Chinnery, P.F.; Surani, M.A. A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell 2015, 161, 1453–1467.
- Kurimoto, K.; Yamaji, M.; Seki, Y.; Saitou, M. Specification of the germ cell lineage in mice: A process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle 2008, 7, 3514–3518.
- Hargan-Calvopina, J.; Taylor, S.; Cook, H.; Hu, Z.; Lee, S.A.; Yen, M.R.; Chiang, Y.S.; Chen, P.Y.; Clark, A.T. Stage-Specific Demethylation in Primordial Germ Cells Safeguards against Precocious Differentiation. Dev. Cell 2016, 39, 75–86.
- Matsui, Y.; Hayashi, K. Epigenetic regulation for the induction of meiosis. Cell. Mol. Life Sci CMLS 2007, 64, 257–262.
- Sachs, M.; Onodera, C.; Blaschke, K.; Ebata, K.T.; Song, J.S.; Ramalho-Santos, M. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 2013, 3, 1777–1784.
- De Felici, M. Nuclear reprogramming in mouse primordial germ cells: Epigenetic contribution. Stem Cells Int. 2011, 2011, 425863.
- Yokobayashi, S.; Liang, C.Y.; Kohler, H.; Nestorov, P.; Liu, Z.; Vidal, M.; van Lohuizen, M.; Roloff, T.C.; Peters, A.H. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature 2013, 495, 236–240.
- Fu, X.F.; Yang, F.; Cheng, S.F.; Feng, Y.N.; Li, L.; Dyce, P.W.; Shen, W.; Sun, X.F. The epigenetic modifications and the anterior to posterior characterization of meiotic entry during mouse oogenesis. Histochem. Cell Biol. 2017, 148, 61–72.
- Kawabata, Y.; Kamio, A.; Jincho, Y.; Sakashita, A.; Takashima, T.; Kobayashi, H.; Matsui, Y.; Kono, T. Sex-specific histone modifications in mouse fetal and neonatal germ cells. Epigenomics 2019, 11, 543–561.
- Mochizuki, K.; Tachibana, M.; Saitou, M.; Tokitake, Y.; Matsui, Y. Implication of DNA Demethylation and Bivalent Histone Modification for Selective Gene Regulation in Mouse Primordial Germ Cells. PLoS ONE 2012, 7, e46036.
- Hackett, J.A.; Sengupta, R.; Zylicz, J.J.; Murakami, K.; Lee, C.; Down, T.A.; Surani, M.A. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 2013, 339, 448–452.
- Hu, Y.-C.; Nicholls, P.K.; Soh, Y.Q.S.; Daniele, J.R.; Junker, J.P.; van Oudenaarden, A.; Page, D.C. Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling. PLOS Genet. 2015, 11, e1005019.
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935.
- Vincent, J.J.; Huang, Y.; Chen, P.Y.; Feng, S.; Calvopiña, J.H.; Nee, K.; Lee, S.A.; Le, T.; Yoon, A.J.; Faull, K.; et al. Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell 2013, 12, 470–478.
- Yamaguchi, S.; Hong, K.; Liu, R.; Shen, L.; Inoue, A.; Diep, D.; Zhang, K.; Zhang, Y. Tet1 controls meiosis by regulating meiotic gene expression. Nature 2012, 492, 443–447.
- Hill, P.W.S.; Leitch, H.G.; Requena, C.E.; Sun, Z.; Amouroux, R.; Roman-Trufero, M.; Borkowska, M.; Terragni, J.; Vaisvila, R.; Linnett, S.; et al. Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature 2018, 555, 392–396.
- Welling, M.; Chen, H.-H.; Muñoz, J.; Musheev, M.U.; Kester, L.; Junker, J.P.; Mischerikow, N.; Arbab, M.; Kuijk, E.; Silberstein, L.; et al. DAZL regulates Tet1 translation in murine embryonic stem cells. EMBO Rep. 2015, 16, 791–802.
- Ito, T.; Osada, A.; Ohta, M.; Yokota, K.; Nishiyama, A.; Niikura, Y.; Tamura, T.; Sekita, Y.; Kimura, T. SWI/SNF chromatin remodeling complex is required for initiation of sex-dependent differentiation in mouse germline. Sci. Rep. 2021, 11, 24074.
- Lawson, K.A.; Dunn, N.R.; Roelen, B.A.; Zeinstra, L.M.; Davis, A.M.; Wright, C.V.; Korving, J.P.; Hogan, B.L. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999, 13, 424–436.
- Ohinata, Y.; Ohta, H.; Shigeta, M.; Yamanaka, K.; Wakayama, T.; Saitou, M. A signaling principle for the specification of the germ cell lineage in mice. Cell 2009, 137, 571–584.
- Saitou, M.; Barton, S.C.; Surani, M.A. A molecular programme for the specification of germ cell fate in mice. Nature 2002, 418, 293–300.
- Ohinata, Y.; Payer, B.; O’Carroll, D.; Ancelin, K.; Ono, Y.; Sano, M.; Barton, S.C.; Obukhanych, T.; Nussenzweig, M.; Tarakhovsky, A.; et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 2005, 436, 207–213.
- Yamaji, M.; Seki, Y.; Kurimoto, K.; Yabuta, Y.; Yuasa, M.; Shigeta, M.; Yamanaka, K.; Ohinata, Y.; Saitou, M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008, 40, 1016–1022.
- De Felici, M.; Pesce, M. Growth factors in mouse primordial germ cell migration and proliferation. Prog. Growth Factor Res. 1994, 5, 135–143.
- De Felici, M. Primordial germ cell biology at the beginning of the XXI century. Int. J. Dev. Biol. 2009, 53, 891–894.
- Ruggiu, M.; Speed, R.; Taggart, M.; McKay, S.J.; Kilanowski, F.; Saunders, P.; Dorin, J.; Cooke, H.J. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 1997, 389, 73–77.
- Schrans-Stassen, B.H.; Saunders, P.T.; Cooke, H.J.; de Rooij, D.G. Nature of the spermatogenic arrest in Dazl −/− mice. Biol. Reprod. 2001, 65, 771–776.
- Gill, M.E.; Hu, Y.C.; Lin, Y.; Page, D.C. Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc. Natl. Acad. Sci. USA 2011, 108, 7443–7448.
- Reijo, R.A.; Dorfman, D.M.; Slee, R.; Renshaw, A.A.; Loughlin, K.R.; Cooke, H.; Page, D.C. DAZ family proteins exist throughout male germ cell development and transit from nucleus to cytoplasm at meiosis in humans and mice. Biol. Reprod. 2000, 63, 1490–1496.
- Sekinaka, T.; Hayashi, Y.; Noce, T.; Niwa, H.; Matsui, Y. Selective de-repression of germ cell-specific genes in mouse embryonic fibroblasts in a permissive epigenetic environment. Sci. Rep. 2016, 6, 32932.
- Zhang, L.; Zhu, R.; Zuo, Q.; Li, D.; Lian, C.; Tang, B.; Xiao, T.; Zhang, Y.; Li, B. Activity analysis and preliminary inducer screening of the chicken DAZL gene promoter. Int. J. Mol. Sci. 2015, 16, 6595–6605.
- Fu, X.-F.; Cheng, S.-F.; Wang, L.-Q.; Yin, S.; De Felici, M.; Shen, W. DAZ Family Proteins, Key Players for Germ Cell Development. Int. J. Biol. Sci. 2015, 11, 1226–1235.
- Collier, B.; Gorgoni, B.; Loveridge, C.; Cooke, H.J.; Gray, N.K. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005, 24, 2656–2666.
- Reynolds, N.; Collier, B.; Maratou, K.; Bingham, V.; Speed, R.M.; Taggart, M.; Semple, C.A.; Gray, N.K.; Cooke, H.J. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum. Mol. Genet. 2005, 14, 3899–3909.
- Lin, Y.; Gill, M.E.; Koubova, J.; Page, D.C. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Science 2008, 322, 1685–1687.
- Soh, Y.Q.S.; Junker, J.P.; Gill, M.E.; Mueller, J.L.; van Oudenaarden, A.; Page, D.C. A Gene Regulatory Program for Meiotic Prophase in the Fetal Ovary. PLOS Genet. 2015, 11, e1005531.
- Wang, Q.; Liu, X.; Tang, N.; Archambeault, D.R.; Li, J.; Song, H.; Tang, C.; He, B.; Matzuk, M.M.; Wang, Y. GASZ promotes germ cell derivation from embryonic stem cells. Stem Cell Res. 2013, 11, 845–860.
- Rosario, R.; Adams, I.R.; Anderson, R.A. Is there a role for DAZL in human female fertility? Mol. Hum. Reprod. 2016, 22, 377–383.
- Jung, D.; Xiong, J.; Ye, M.; Qin, X.; Li, L.; Cheng, S.; Luo, M.; Peng, J.; Dong, J.; Tang, F.; et al. In vitro differentiation of human embryonic stem cells into ovarian follicle-like cells. Nat. Commun. 2017, 8, 15680.
- Rosario, R.; Smith, R.W.; Adams, I.R.; Anderson, R.A. RNA immunoprecipitation identifies novel targets of DAZL in human foetal ovary. Mol. Hum. Reprod. 2017, 23, 177–186.
- Toyooka, Y.; Tsunekawa, N.; Takahashi, Y.; Matsui, Y.; Satoh, M.; Noce, T. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech. Dev. 2000, 93, 139–149.
- Lasko, P. The DEAD-box helicase Vasa: Evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2013, 1829, 810–816.
- Xu, C.; Cao, Y.; Bao, J. Building RNA-protein germ granules: Insights from the multifaceted functions of DEAD-box helicase Vasa/Ddx4 in germline development. Cell. Mol. Life Sci. 2021, 79, 4.
- Tanaka, S.S.; Toyooka, Y.; Akasu, R.; Katoh-Fukui, Y.; Nakahara, Y.; Suzuki, R.; Yokoyama, M.; Noce, T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000, 14, 841–853.
- De Miguel, M.P.; Alcaina, Y.; de la Maza, D.S. Primordial germ cell reprogramming. In Germ Cell; IntechOpen: London, UK, 2018; pp. 43–62.
- Donovan, P.J.; de Miguel, M.P. Turning germ cells into stem cells. Curr. Opin. Genet. Dev. 2003, 13, 463–471.
- Overeem, A.W.; Chang, Y.W.; Spruit, J.; Roelse, C.M.; Chuva De Sousa Lopes, S.M. Ligand-Receptor Interactions Elucidate Sex-Specific Pathways in the Trajectory from Primordial Germ Cells to Gonia During Human Development. Front. Cell Dev. Biol. 2021, 9, 661243.
- Western, P.S.; Miles, D.C.; van den Bergen, J.A.; Burton, M.; Sinclair, A.H. Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells 2008, 26, 339–347.
- Spiller, C.; Wilhelm, D.; Koopman, P. Cell cycle analysis of fetal germ cells during sex differentiation in mice. Biol. Cell 2009, 101, 587–598.
- Watanabe, Y.; Yokobayashi, S.; Yamamoto, M.; Nurse, P. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature 2001, 409, 359–363.
- Forsburg, S.L.; Hodson, J.A. Mitotic replication initiation proteins are not required for pre-meiotic S phase. Nat. Genet. 2000, 25, 263–268.
- Murakami, H.; Nurse, P. Regulation of premeiotic S phase and recombination-related double-strand DNA breaks during meiosis in fission yeast. Nat. Genet. 2001, 28, 290–293.
- Callan, H.G. Replication of DNA in the chromosomes of eukaryotes. Proc. R. Soc. London. Ser. B Biol. Sci. 1972, 181, 19–41.
- Lee, B.; Amon, A. Meiosis: How to create a specialized cell cycle. Curr. Opin. Cell Biol. 2001, 13, 770–777.
- Williamson, D.H.; Johnston, L.H.; Fennell, D.J.; Simchen, G. The timing of the S phase and other nuclear events in yeast meiosis. Exp. Cell Res. 1983, 145, 209–217.
- Ishiguro, K.I.; Matsuura, K.; Tani, N.; Takeda, N.; Usuki, S.; Yamane, M.; Sugimoto, M.; Fujimura, S.; Hosokawa, M.; Chuma, S.; et al. MEIOSIN Directs the Switch from Mitosis to Meiosis in Mammalian Germ Cells. Dev. Cell 2020, 52, 429–445.e10.
- Kojima, M.L.; de Rooij, D.G.; Page, D.C. Amplification of a broad transcriptional program by a common factor triggers the meiotic cell cycle in mice. eLife 2019, 8, e43738.
- Kumar, S.; Chatzi, C.; Brade, T.; Cunningham, T.J.; Zhao, X.; Duester, G. Sex-specific timing of meiotic initiation is regulated by Cyp26b1 independent of retinoic acid signalling. Nat. Commun. 2011, 2, 151.
- Vernet, N.; Condrea, D.; Mayere, C.; Féret, B.; Klopfenstein, M.; Magnant, W.; Alunni, V.; Teletin, M.; Souali-Crespo, S.; Nef, S.; et al. Meiosis occurs normally in the fetal ovary of mice lacking all retinoic acid receptors. Sci. Adv. 2020, 6, eaaz1139.
- Chassot, A.A.; Le Rolle, M.; Jolivet, G.; Stevant, I.; Guigonis, J.M.; Da Silva, F.; Nef, S.; Pailhoux, E.; Schedl, A.; Ghyselinck, N.B.; et al. Retinoic acid synthesis by ALDH1A proteins is dispensable for meiosis initiation in the mouse fetal ovary. Sci. Adv. 2020, 6, eaaz1261.
- Bellutti, L.; Abby, E.; Tourpin, S.; Messiaen, S.; Moison, D.; Trautmann, E.; Guerquin, M.-J.; Rouiller-Fabre, V.; Habert, R.; Livera, G. Divergent Roles of CYP26B1 and Endogenous Retinoic Acid in Mouse Fetal Gonads. Biomolecules 2019, 9, 536.
- Endo, T.; Mikedis, M.M.; Nicholls, P.K.; Page, D.C.; de Rooij, D.G. Retinoic Acid and Germ Cell Development in the Ovary and Testis. Biomolecules 2019, 9, 775.
- Duester, G. Retinoic acid synthesis and signaling during early organogenesis. Cell 2008, 134, 921–931.
- Bowles, J.; Knight, D.; Smith, C.; Wilhelm, D.; Richman, J.; Mamiya, S.; Yashiro, K.; Chawengsaksophak, K.; Wilson, M.J.; Rossant, J.; et al. Retinoid signaling determines germ cell fate in mice. Science 2006, 312, 596–600.
- Mu, X.; Wen, J.; Guo, M.; Wang, J.; Li, G.; Wang, Z.; Wang, Y.; Teng, Z.; Cui, Y.; Xia, G. Retinoic acid derived from the fetal ovary initiates meiosis in mouse germ cells. J. Cell. Physiol. 2013, 228, 627–639.
- Bowles, J.; Feng, C.W.; Miles, K.; Ineson, J.; Spiller, C.; Koopman, P. ALDH1A1 provides a source of meiosis-inducing retinoic acid in mouse fetal ovaries. Nat. Commun. 2016, 7, 10845.
- Bullejos, M.; Koopman, P. Germ cells enter meiosis in a rostro-caudal wave during development of the mouse ovary. Mol. Reprod. Dev. 2004, 68, 422–428.
- Menke, D.B.; Koubova, J.; Page, D.C. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev. Biol. 2003, 262, 303–312.
- Giuili, G.; Tomljenovic, A.; Labrecque, N.; Oulad-Abdelghani, M.; Rassoulzadegan, M.; Cuzin, F. Murine spermatogonial stem cells: Targeted transgene expression and purification in an active state. EMBO Rep. 2002, 3, 753–759.
- Feng, C.-W.; Burnet, G.; Spiller, C.M.; Cheung, F.K.M.; Chawengsaksophak, K.; Koopman, P.; Bowles, J. Identification of regulatory elements required for Stra8 expression in fetal ovarian germ cells of the mouse. Development 2021, 148, dev194977.
- Oulad-Abdelghani, M.; Bouillet, P.; Décimo, D.; Gansmuller, A.; Heyberger, S.; Dollé, P.; Bronner, S.; Lutz, Y.; Chambon, P. Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J. Cell Biol. 1996, 135, 469–477.
- Baltus, A.E.; Menke, D.B.; Hu, Y.-C.; Goodheart, M.L.; Carpenter, A.E.; de Rooij, D.G.; Page, D.C. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat. Genet. 2006, 38, 1430–1434.
- Koubova, J.; Menke, D.B.; Zhou, Q.; Capel, B.; Griswold, M.D.; Page, D.C. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc. Natl. Acad. Sci. USA 2006, 103, 2474–2479.
- Anderson, E.L.; Baltus, A.E.; Roepers-Gajadien, H.L.; Hassold, T.J.; de Rooij, D.G.; van Pelt, A.M.M.; Page, D.C. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc. Natl. Acad. Sci. USA 2008, 105, 14976–14980.
- Mark, M.; Jacobs, H.; Oulad-Abdelghani, M.; Dennefeld, C.; Féret, B.; Vernet, N.; Codreanu, C.A.; Chambon, P.; Ghyselinck, N.B. STRA8-deficient spermatocytes initiate, but fail to complete, meiosis and undergo premature chromosome condensation. J. Cell Sci. 2008, 121, 3233–3242.
- Massari, M.E.; Murre, C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000, 20, 429–440.
- Bowles, J.; Feng, C.W.; Spiller, C.; Davidson, T.L.; Jackson, A.; Koopman, P. FGF9 suppresses meiosis and promotes male germ cell fate in mice. Dev. Cell 2010, 19, 440–449.
- Barrios, F.; Filipponi, D.; Pellegrini, M.; Paronetto, M.P.; Di Siena, S.; Geremia, R.; Rossi, P.; De Felici, M.; Jannini, E.A.; Dolci, S. Opposing effects of retinoic acid and FGF9 on Nanos2 expression and meiotic entry of mouse germ cells. J. Cell Sci. 2010, 123, 871–880.
- Feng, Y.M.; Liang, G.J.; Pan, B.; Qin, X.S.; Zhang, X.F.; Chen, C.L.; Li, L.; Cheng, S.F.; De Felici, M.; Shen, W. Notch pathway regulates female germ cell meiosis progression and early oogenesis events in fetal mouse. Cell Cycle 2014, 13, 782–791.
- Souquet, B.; Tourpin, S.; Messiaen, S.; Moison, D.; Habert, R.; Livera, G. Nodal Signaling Regulates the Entry into Meiosis in Fetal Germ Cells. Endocrinology 2012, 153, 2466–2473.
- De Bellis, M.; Carbonara, R.; Roussel, J.; Farinato, A.; Massari, A.; Pierno, S.; Muraglia, M.; Corbo, F.; Franchini, C.; Carratu, M.R.; et al. Increased sodium channel use-dependent inhibition by a new potent analogue of tocainide greatly enhances in vivo antimyotonic activity. Neuropharmacology 2017, 113, 206–216.
- Le Rolle, M.; Massa, F.; Siggers, P.; Turchi, L.; Loubat, A.; Koo, B.K.; Clevers, H.; Greenfield, A.; Schedl, A.; Chaboissier, M.C.; et al. Arrest of WNT/β-catenin signaling enables the transition from pluripotent to differentiated germ cells in mouse ovaries. Proc. Natl. Acad. Sci. USA 2021, 118, e2023376118.
- Liang, G.J.; Zhang, X.F.; Wang, J.J.; Sun, Y.C.; Sun, X.F.; Cheng, S.F.; Li, L.; De Felici, M.; Shen, W. Activin A accelerates the progression of fetal oocytes throughout meiosis and early oogenesis in the mouse. Stem Cells Dev. 2015, 24, 2455–2465.
- Zhang, X.; Gunewardena, S.; Wang, N. Nutrient restriction synergizes with retinoic acid to induce mammalian meiotic initiation in vitro. Nat. Commun. 2021, 12, 1758.
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174.
- Matsuhara, H.; Yamamoto, A. Autophagy is required for efficient meiosis progression and proper meiotic chromosome segregation in fission yeast. Genes Cells Devoted Mol. Cell. Mech. 2016, 21, 65–87.
- Wen, F.P.; Guo, Y.S.; Hu, Y.; Liu, W.X.; Wang, Q.; Wang, Y.T.; Yu, H.Y.; Tang, C.M.; Yang, J.; Zhou, T.; et al. Distinct temporal requirements for autophagy and the proteasome in yeast meiosis. Autophagy 2016, 12, 671–688.
- Ferder, I.C.; Fung, L.; Ohguchi, Y.; Zhang, X.; Lassen, K.G.; Capen, D.; Brown, D.; Xavier, R.J.; Wang, N. Meiotic gatekeeper STRA8 suppresses autophagy by repressing Nr1d1 expression during spermatogenesis in mice. PLoS Genet. 2019, 15, e1008084.
- Zheng, X.-F.; Schreiber, S.L. Target of rapamycin proteins and their kinase activities are required for meiosis. Proc. Natl. Acad. Sci. USA 1997, 94, 3070–3075.
- Wei, Y.; Reveal, B.; Reich, J.; Laursen, W.J.; Senger, S.; Akbar, T.; Iida-Jones, T.; Cai, W.; Jarnik, M.; Lilly, M.A. TORC1 regulators Iml1/GATOR1 and GATOR2 control meiotic entry and oocyte development in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E5670–E5677.
- Shimada, R.; Koike, H.; Hirano, T.; Saga, Y. NANOS2 suppresses the cell cycle by repressing mTORC1 activators in embryonic male germ cells. bioRxiv 2020.




