
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Haigang Ren | + 1543 word(s) | 1543 | 2020-10-07 20:14:03 | | | |
| 2 | Nicole Yin | Meta information modification | 1543 | 2020-10-20 10:23:25 | | |
Video Upload Options
Acetylation of lysine residues is a key post-translational modification for protein functions in all eukaryotic organisms. Acetylation of lysine residues can be catalyzed by lysine acetyltransferases (KATs) or modified by abundant Ac-CoA through nonenzymatic mechanisms. Conversely, lysine deacetylation is catalyzed by lysine deacetylases (KDACs).
1. Introduction
Acetylation of lysine residues originally discovered in 1964 as a unique post-translational modification of histones, modifications of lysine acetylation and deacetylation are now found in thousands of nonhistone proteins which are located in virtually every cellular compartment and have essential roles in various cellular processes including gene regulation, cell signaling, and metabolism, as well as contribute to the progression of multiple diseases.
2. Lysine Acetylation and Its Regulatory Mechanism and Functions
2.1. Lysine Acetylation and Its Functions
Lysine acetylation in histones was first described by Vincent Allfrey and his colleagues in 1964[1]. Lysine acetylation is an evolutionarily conserved and reversible posttranslational modification (PTM) in eukaryotes that precisely governs protein functions and involves transfer of an acetyl group donated by acetyl coenzyme A (Ac-CoA) to the ε-amino side chain of a protein lysine residue. Lysine acetylation occurs in both histones and nonhistone proteins. Lysine acetylation of histones such as Histone 2A (H2A), Histone 2B (H2B), Histone 3 (H3), and Histone 4 (H4) generally results in transcriptional activation due to destabilization of DNA-histone binding, as acetylation of lysine neutralizes its positive charge, which prevents the formation of salt bridges with the negatively charged phosphate backbone of DNA[2]. In addition to histones, many nonhistone proteins in the cytoplasm and organelles are also dynamically acetylated and deacetylated; these changes are closely implicated in the regulation of various cellular processes, including gene transcription; cell cycle progression; DNA damage repair; cellular signal transduction; protein folding stability and aggregation; cytoskeleton organization; RNA processing and stability; and autophagy regulation[3][4].
Acetylation of lysine residues can be catalyzed by KATs or modified by abundant Ac-CoA through nonenzymatic mechanisms. Conversely, lysine deacetylation is catalyzed by KDACs, which comprise two major groups with distinct catalytic mechanisms: NAD+-dependent Sirtuins (SIRTs) and Zn2+-dependent histone deacetylases (HDACs) (Figure 1). The acetylation levels of lysines are highly dynamic, and the balance between lysine acetylation and deacetylation is precisely controlled by KATs and KDACs as well as by the concentration of Ac-CoA in organellar compartments such as mitochondria[5][6].
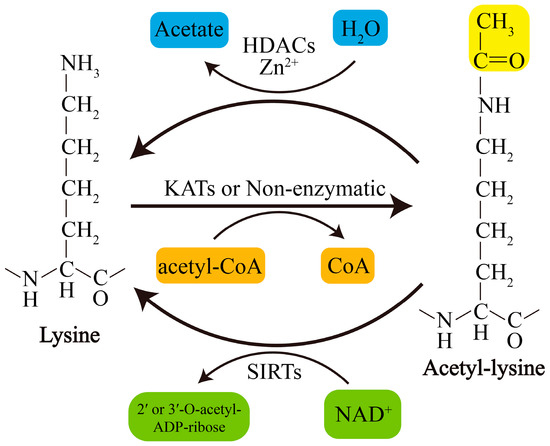
Figure 1. Schematic overview of lysine acetylation and deacetylation. Lysine acetylation, which is catalyzed by KATs, involves transfer of an acetyl group from Ac-CoA to the ε-amino side chain of lysine or occurs nonenzymatically. Deacetylation of lysine residues is catalyzed by Zn2+-dependent HDACs or by NAD+-dependent SIRTs. NAD+, nicotinamide adenine dinucleotide.
2.2. KATs and KDACs in Humans and Their Involvement in PD
To date, at least 22 human KATs have been identified to display acetyltransferase activity; these KATs can be divided into three major families: the MYST family, the GNAT family, and the p300/CBP family[4][7] (Table 1). The substrate specificity of KATs is primarily determined by their subcellular distribution or interacting partners or by the availability of lysine in substrates[3]. Most KATs are localized mainly in the nucleus, where they mediate processes including but not limited to histone acetylation, and some KATs also located in the cytoplasm are responsible for cytoplasmic substrate acetylation[3]. Recently, GCN5-like 1 (GCN5L1) and Ac-CoA Acetyltransferase 1 (ACAT1) were identified as mitochondrial KATs that regulate mitochondrial functions by acetylating several mitochondrial substrates[8][9]. In addition, the well-known nuclear KAT8/MOF is also found to localize to mitochondria and affect mitochondrial functions[10]. The classifications, subcellular localization, involvement in PD models, and relevant substrates of these KATs are presented in Table 1. However, to date, only a small proportion of KATs have been identified to be related to PD (Table 1).
Table 1. Human KATs and their involvement in PD.
|
Family |
Name |
Subcellular Localization |
PD Model/KAT Change/Substrate Acetylation Changes |
|
GNAT |
KAT1/HAT1 |
Nucleus |
Mn/expression ↓/H3 and H4 ↓ [11]. |
|
GNAT |
KAT2A/GCN5 |
Nucleus |
MPP+/activity ↑/PGC-1α ↑ [12]. |
|
GNAT |
KAT2B/PCAF |
Nucleus |
NA |
|
GNAT |
KAT9/ELP3 |
Nucleus/cytoplasm |
NA |
|
GNAT |
αTAT1/ATAT1 |
Cytoplasm/membrane |
LRRK2 knockout/NA/α-tubulin ↑; LRKK2 R1441C or Y1699C/NA/α-tubulin ↓[13][14]. |
|
p300/CBP |
KAT3A/CBP |
Nucleus/cytoplasm |
Dieldrin/expression ↑/H3 and H4 ↑[15]. |
|
p300/CBP |
KAT3B/p300 |
Nucleus/cytoplasm |
|
|
MYST |
KAT5/TIP60/PLIP |
Nucleus/cytoplasm |
NA |
|
MYST |
KAT6A/MOZ/MYST3 |
Nucleus |
NA |
|
MYST |
KAT6A/MORF/MYST4 |
Nucleus |
NA |
|
MYST |
KAT7/HBO1/MYST2 |
Nucleus |
NA |
|
MYST |
KAT8/MOF/MYST1 |
Nucleus/mitochondria |
NA |
|
Other |
KAT4/TAF1/TAFII250 |
Nucleus |
NA |
|
Other |
KAT12/TFIIC90 |
Nucleus |
NA |
|
Other |
KAT13A/SRC-1/NCOA1 |
Nucleus |
NA |
|
Other |
KAT13B/SRC-3/NCOA3 |
Nucleus/cytoplasm |
NA |
|
Other |
KAT13C/SRC-2/NCOA2 |
Nucleus |
NA |
|
Other |
KAT13D/CLOCK |
Nucleus/cytoplasm |
NA |
|
Other |
ATF-2/CREB2 |
Nucleus/cytoplasm |
NA |
|
Other |
NAT10 |
Nucleus |
NA |
|
Other |
ACAT1 |
Mitochondria |
NA |
|
Other |
GCN5L1 |
Mitochondria |
NA |
↑, upregulation; ↓, downregulated; NA, not available; PGC-1α, peroxisome proliferator-activated receptor γ coactivator-1α; NF-κB, nuclear factor Kappa-B.
KDACs, originally referred to HDACs, were initially discovered to deacetylate histones in 1995[18]. Later, they were also found to regulate nonhistone protein acetylation and cellular functions[19]. Currently, KDACs are grouped into two major types: NAD+-dependent SIRTs (SIRT1-7) and Zn2+-dependent HDACs (HDAC1-11). They can also be divided into four categories according to phylogeny and sequence similarities: Class I, Class IIa, Class IIb, and Class IV (Table 2). Recently, lymphoid enhancer-binding factor 1 (LEF1) and T cell-specific transcription factor 1 (TCF1) were identified as novel KDACs that are not related to the abovementioned types of KDACs[20]. Zn2+-dependent HDACs are primarily distributed in the nucleus or cytoplasm, although HDAC1 and HDAC7 are also found in mitochondria in some types of cells or under certain conditions[21][22]. In contrast, some SIRTs, including SIRT3-5, are restricted to the mitochondria, indicating their unique and crucial roles in mitochondria. However, it should be noted that several KDACs, such as that of SIRT4-6 and some class IIa HDACs, display weak or no deacetylase activity or target other types of acylation[3]. For example, SIRT5 exerts the activity of desuccinylase, demalonylase, and deglutarylase[23]; SIRT4 removes the acyl moieties from lysine residues such as methylglutaryl-, hydroxymethylglutaryl- and 3-methylglutaconyl-lysine[24]; SIRT6 functions to deacetylate long-chain fatty acyl groups rather than protein deacetylation[25]; The classifications, subcellular localization of KDACs, as well as their involvement in PD and the relevant acetylation of substrates are presented in Table 2.
Interestingly, the activity or expression levels of nuclear SIRT1 and mitochondrial SIRT3 are consistently decreased in PD tissues and different PD models. The activity or expression levels of nuclear HDAC2 and HDAC3 are increased in most PD models, but the expression levels of HDAC2 are decreased in tissues of PD patients. Furthermore, the activity or expression levels of two main cytoplasmic KDACs, HDAC6 and SIRT2, are downregulated and upregulated in most PD models, respectively (Table 3). Of note, beyond acetylation, several KATs/KDACs have activity of other acylation modifications including propionyl, butyryl, 2-hydroxyisobutyryl, crotonyl, malonyl, succinyl, or glutaryl modification. For example, p300 has crotonyltransferase activity[26], while KAT2A/GCN5 has both crotonyltransferase and uccinyltransferase activity[27][28], whereas HDAC1/2/3/8 and SIRT1-3 possess decrotonylating activity[29][30][31]. Whether these changes in KATs or KDACs in PD patients or models also cause variation of other acylation modifications, and the roles of these acylation variations in PD pathology, deserve further research.
Table 2. Human KDACs and their involvement in PD.
|
Class |
Name |
Subcellular Localization |
PD Model/KDAC Change/Substrate Acetylation Level Change |
|
I |
HDAC1 |
Nucleus |
Patient tissues, MPTP or MPP+/expression ↓/H2A, H2B, H3 and H4 ↑[32]. |
|
I |
HDAC2 |
Nucleus |
Patient tissues, MPTP or MPP+/expression ↓/H2A, H2B, H3 and H4 ↑[32]; MPP+/expression ↑/NA[33]; idiopathic PD fibroblasts/expression ↑/H3 ↓; LRRK2 G2109S PD fibroblasts/expression ↑/H3 ↓ [34]. |
|
I |
HDAC3 |
Nucleus |
Idiopathic PD fibroblasts/expression ↑/H3 ↓ [34]; Mn/expression ↑/H3 and H4 ↓ [11]; LRRK2 or mutation/phosphorylation ↑, nuclear translocation ↑ and activity ↑/H4 ↓ [35]; PINK1 mutation/phosphorylation ↓ and activity ↓/p53 ↑[36]. |
|
I |
HDAC8 |
Nucleus/cytoplasm |
NA |
|
IIa |
HDAC4 |
Nucleus/cytoplasm |
Patient tissues, MPTP or MPP+/expression ↓/H2A, H2B, H3 and H4 ↑[32]; paraquat/expression ↓/H3 ↑[37][38]; idiopathic PD fibroblasts/expression ↑/H3 ↓, LRRK2 G2109S PD fibroblasts/expression ↑/H3 ↓ [48]; Mn/expression ↑/H3 and H4 ↓ [11]. |
|
IIa |
HDAC5 |
Nucleus/cytoplasm |
NA |
|
IIa |
HDAC7 |
Nucleus/cytoplasm |
|
|
IIa |
HDAC9 |
Nucleus/cytoplasm |
NA |
|
IIb |
HDAC6 |
Primarily cytoplasm |
Patient tissues, MPTP or MPP+/expression ↓[32]; idiopathic PD fibroblasts/expression ↓, LRRK2 G2109S PD fibroblasts/expression ↓ [34]; Parkin absence/NA/α-tubulin ↑ [39]; ATP13A absence/activity ↓/α-tubulin ↑[40]; 6-OHDA/expression ↑/peroxiredoxin 1/2 ↓ [41]. |
|
IIb |
HDAC10 |
Primarily cytoplasm |
NA |
|
III |
SIRT1 |
Nucleus |
Patient tissues, MPTP or MPP+/expression ↓/H2A, H2B, H3 and H4 ↑ [32]; patient tissues/activity ↓ [42]; MPTP/expression ↓/LC3 ↑ [43]; MPTP/S-nitrosylation ↑ and activity ↓/p53 and NFκB-p65 ↑ [44]; MPP+/expression ↓/H3 and PGC-1α ↑[45]; rotenone/expression ↓/H3 ↑[46][47]; 6-OHDA/expression ↓/BMAL1 ↑ [48]; LRRK2 G2019S iPSC-derived dopaminergic cultures/activity ↓/p53 ↑ [49]. |
|
III |
SIRT2 |
Cytoplasm |
MPTP/activity ↑/α-syn ↓[50] [64]; α-syn/activity ↑/α-tubulin ↓[51]; MPTP or MPP+/activity ↑/Foxo3a ↓ [52]; 6-OHDA/activity ↓/α-tubulin ↑[53]. |
|
III |
SIRT3 |
Mitochondria |
Patients/NA/MnSOD ↑[54]; MPTP/expression ↓/SOD2 and ATP5B ↑[55]; MPP+/expression ↓/citrate synthase and isocitrate dehydrogenase 2 ↑[56]; α-syn/expression ↓/SOD2 ↑ [57]; LRRK2 G2019S iPSC-derived dopaminergic cultures/activity ↓/SOD2 ↑ [49]. |
|
III |
SIRT4 |
Mitochondria |
NA |
|
III |
SIRT5 |
Mitochondria |
NA |
|
III |
SIRT6 |
Nucleus |
NA |
|
III |
SIRT7 |
Nucleolus |
NA |
|
IV |
HDAC11 |
Primarily nucleus |
NA |
|
Other |
TCF1 |
Nucleus |
NA |
|
Other |
LEF1 |
Nucleus |
NA |
↑, upregulation; ↓, downregulated; NA, not available, Mn, manganese; BMAL1, brain and muscle arnt-like 1; iPSC, induced pluripotent stem cells, Foxo3a, Forkhead box O3; ATP5B, ATP synthase subunit β; SOD2, superoxide dismutase.
References
- V. G. Allfrey; R. Faulkner; A. E. Mirsky; ACETYLATION AND METHYLATION OF HISTONES AND THEIR POSSIBLE ROLE IN THE REGULATION OF RNA SYNTHESIS. Proceedings of the National Academy of Sciences 1964, 51, 786-794, 10.1073/pnas.51.5.786.
- Peter Tessarz; Tony Kouzarides; Histone core modifications regulating nucleosome structure and dynamics. Nature Reviews Molecular Cell Biology 2014, 15, 703-708, 10.1038/nrm3890.
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174.
- Sheikh, B.N.; Akhtar, A. The many lives of KATs - detectors, integrators and modulators of the cellular environment. Nat. Rev. Genet. 2019, 20, 7–23.
- Hosp, F.; Lassowskat, I.; Santoro, V.; De Vleesschauwer, D.; Fliegner, D.; Redestig, H.; Mann, M.; Christian, S.; Hannah, M.A.; Finkemeier, I. Lysine acetylation in mitochondria: From inventory to function. Mitochondrion 2017, 33, 58–71.
- Li, P.; Ge, J.; Li, H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat. Rev. Cardiol. 2020, 17, 96–115.
- Adrian Drazic; Line M. Myklebust; Rasmus Ree; Thomas Arnesen; The world of protein acetylation. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2016, 1864, 1372-1401, 10.1016/j.bbapap.2016.06.007.
- Scott, I.; Webster, B.R.; Li, J.H.; Sack, M.N. Identification of a molecular component of the mitochondrial acetyltransferase programme: A novel role for GCN5L1. Biochem. J. 2012, 443, 655–661.
- Fan, J.; Shan, C.; Kang, H.B.; Elf, S.; Xie, J.; Tucker, M.; Gu, T.L.; Aguiar, M.; Lonning, S.; Chen, H.; et al. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol. Cell 2014, 53, 534–548.
- Aindrila Chatterjee; Janine Seyfferth; Jacopo Lucci; Ralf Gilsbach; Sebastian Preissl; Lena Böttinger; Christoph U. Mårtensson; Amol Panhale; Thomas Stehle; Oliver Kretz; et al.Abdullah H. SahyounSergiy AvilovStefan EimerLutz HeinNikolaus PfannerThomas BeckerAsifa Akhtar MOF Acetyl Transferase Regulates Transcription and Respiration in Mitochondria. Cell 2016, 167, 722-738, 10.1016/j.cell.2016.09.052.
- Zhenkun Guo; Zhipeng Zhang; Qingqing Wang; Jie Zhang; Lijin Wang; Qunwei Zhang; Huangyuan Li; Siying Wu; Manganese chloride induces histone acetylation changes in neuronal cells: Its role in manganese-induced damage. NeuroToxicology 2018, 65, 255-263, 10.1016/j.neuro.2017.11.003.
- Fei Fan; Songlin Li; Zhipeng Wen; Qiaoyue Ye; Xiaochun Chen; Qinyong Ye; Regulation of PGC-1α mediated by acetylation and phosphorylation in MPP+ induced cell model of Parkinson’s disease. Aging 2020, 12, 9461-9474, 10.18632/aging.103219.
- Godena, V.K.; Brookes-Hocking, N.; Moller, A.; Shaw, G.; Oswald, M.; Sancho, R.M.; Miller, C.C.; Whitworth, A.J.; De Vos, K.J. Increasing microtubule acetylation rescues axonal transport and locomotor deficits caused by LRRK2 Roc-COR domain mutations. Nat. Commun. 2014, 5, 5245.
- Law, B.M.; Spain, V.A.; Leinster, V.H.; Chia, R.; Beilina, A.; Cho, H.J.; Taymans, J.M.; Urban, M.K.; Sancho, R.M.; Blanca Ramirez, M.; et al. A direct interaction between leucine-rich repeat kinase 2 and specific beta-tubulin isoforms regulates tubulin acetylation. J. Biol. Chem. 2014, 289, 895–908
- Anumantha Kanthasamy; Huajun Jin; Vellareddy Anantharam; Gautam Sondarva; Velusamy Rangasamy; Ajay Rana; Arthi Kanthasamy; Emerging neurotoxic mechanisms in environmental factors-induced neurodegeneration. NeuroToxicology 2012, 33, 833-837, 10.1016/j.neuro.2012.01.011.
- Jin, H.; Kanthasamy, A.; Ghosh, A.; Yang, Y.; Anantharam, V.; Kanthasamy, A.G. alpha-Synuclein negatively regulates protein kinase Cdelta expression to suppress apoptosis in dopaminergic neurons by reducing p300 histone acetyltransferase activity. J. Neurosci. 2011, 31, 2035–2051.
- Paiva, I.; Pinho, R.; Pavlou, M.A.; Hennion, M.; Wales, P.; Schutz, A.L.; Rajput, A.; Szego, E.M.; Kerimoglu, C.; Gerhardt, E.; et al. Sodium butyrate rescues dopaminergic cells from alpha-synuclein-induced transcriptional deregulation and DNA damage. Hum. Mol. Genet. 2017, 26, 2231–2246.
- Susanne Kleff; Erik D. Andrulis; Carl W. Anderson; Rolf Sternglanz; Identification of a Gene Encoding a Yeast Histone H4 Acetyltransferase. Journal of Biological Chemistry 1995, 270, 24674-24677, 10.1074/jbc.270.42.24674.
- Xiang-Jiao Yang; Edward Seto; Lysine Acetylation: Codified Crosstalk with Other Posttranslational Modifications. Molecular Cell 2008, 31, 449-461, 10.1016/j.molcel.2008.07.002.
- Shaojun Xing; Fengyin Li; Zhouhao Zeng; Yunjie Zhao; Shuyang Yu; Qiang Shan; Yalan Li; Farrah C. Phillips; Peterson K. Maina; Hank H. Qi; et al.Chengyu LiuJun ZhuR. Marshall PopeCatherine A. MusselmanChen ZengWeiqun PengHai-Hui “Howard” Xue Tcf1 and Lef1 transcription factors establish CD8+ T cell identity through intrinsic HDAC activity. Nature Immunology 2016, 17, 695-703, 10.1038/ni.3456.
- Herr, D.J.; Baarine, M.; Aune, S.E.; Li, X.; Ball, L.E.; Lemasters, J.J.; Beeson, C.C.; Chou, J.C.; Menick, D.R. HDAC1 localizes to the mitochondria of cardiac myocytes and contributes to early cardiac reperfusion injury. J. Mol. Cell. Cardiol. 2018, 114, 309–319.
- Bakin, R.E.; Jung, M.O. Cytoplasmic sequestration of HDAC7 from mitochondrial and nuclear compartments upon initiation of apoptosis. J. Biol. Chem. 2004, 279, 51218–51225.
- Surinder Kumar; David B. Lombard; Functions of the sirtuin deacylase SIRT5 in normal physiology and pathobiology. Critical Reviews in Biochemistry and Molecular Biology 2018, 53, 311-334, 10.1080/10409238.2018.1458071.
- Kristin A. Anderson; Frank K. Huynh; Kelsey Fisher-Wellman; J. Darren Stuart; Brett S. Peterson; Jonathan D. Douros; Gregory R. Wagner; J. Will Thompson; Andreas S. Madsen; Michelle F. Green; et al.R. Michael SivleyOlga R. IlkayevaRobert D. StevensDonald S. BackosJohn A. CapraChristian A. OlsenJonathan E. CampbellDeborah M. MuoioPaul A. GrimsrudMatthew D. Hirschey SIRT4 Is a Lysine Deacylase that Controls Leucine Metabolism and Insulin Secretion. Cell Metabolism 2017, 25, 838-855, 10.1016/j.cmet.2017.03.003.
- Hong Jiang; Saba Khan; Yi Wang; Guillaume Charron; Bin He; Carlos Sebastian; Jintang Du; Ray Kim; Eva Ge; Raul Mostoslavsky; et al.Howard C. HangQuan HaoHening Lin SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013, 496, 110-113, 10.1038/nature12038.
- Benjamin R. Sabari; Zhanyun Tang; He Huang; Vladimir Yong-Gonzalez; Henrik Molina; Ha Eun Kong; Lunzhi Dai; Miho Shimada; Justin R. Cross; Yingming Zhao; et al.Robert G. RoederC. David Allis Intracellular Crotonyl-CoA Stimulates Transcription through p300-Catalyzed Histone Crotonylation. Molecular Cell 2015, 58, 203-215, 10.1016/j.molcel.2015.02.029.
- Wang, Y.; Jin, J.; Chung, M.W.H.; Feng, L.; Sun, H.; Hao, Q. Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation. Proc. Natl. Acad. Sci. USA 2018, 115, 2365–2370.
- Kollenstart, L.; de Groot, A.J.L.; Janssen, G.M.C.; Cheng, X.; Vreeken, K.; Martino, F.; Cote, J.; van Veelen, P.A.; van Attikum, H. Gcn5 and Esa1 function as histone crotonyltransferases to regulate crotonylation-dependent transcription. J. Biol. Chem. 2019, 294, 20122–20134.
- Feldman, J.L.; Baeza, J.; Denu, J.M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 2013, 288, 31350–31356.
- Wei, W.; Liu, X.; Chen, J.; Gao, S.; Lu, L.; Zhang, H.; Ding, G.; Wang, Z.; Chen, Z.; Shi, T.; et al. Class I histone deacetylases are major histone decrotonylases: Evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017, 27, 898–915.
- Kelly, R.D.W.; Chandru, A.; Watson, P.J.; Song, Y.; Blades, M.; Robertson, N.S.; Jamieson, A.G.; Schwabe, J.W.R.; Cowley, S.M. Histone deacetylase (HDAC) 1 and 2 complexes regulate both histone acetylation and crotonylation in vivo. Sci. Rep. 2018, 8, 14690.
- Goonho Park; Jieqiong Tan; Guillermina Garcia; Yunyi Kang; Guy S Salvesen; Zhuohua Zhang; Regulation of Histone Acetylation by Autophagy in Parkinson Disease. Journal of Biological Chemistry 2015, 291, 3531-3540, 10.1074/jbc.m115.675488.
- Chi-Jing Choong; Tsutomu Sasaki; Hideki Hayakawa; Toru Yasuda; Kousuke Baba; Yoshiyuki Hirata; Shinichi Uesato; Hideki Mochizuki; A novel histone deacetylase 1 and 2 isoform-specific inhibitor alleviates experimental Parkinson's disease. Neurobiology of Aging 2016, 37, 103-116, 10.1016/j.neurobiolaging.2015.10.001.
- Sokhna M. S. Yakhine-Diop; Mireia Niso-Santano; Mario Rodríguez-Arribas; Rubén Gómez-Sánchez; Guadalupe Martínez-Chacón; Elisabet Uribe-Carretero; José A. Navarro-García; Gema Ruiz-Hurtado; Ana Aiastui; J. Mark Cooper; et al.Adolfo López De MunaínJosé M. Bravo-San PedroRosa A. González-PoloJosé M. Fuentes Impaired Mitophagy and Protein Acetylation Levels in Fibroblasts from Parkinson’s Disease Patients. Molecular Neurobiology 2018, 56, 2466-2481, 10.1007/s12035-018-1206-6.
- Kyung Ah Han; Woo Hyun Shin; Sungyeon Jung; Wongi Seol; H. Seo; Chemyong Ko; Kwang Chul Chung; Leucine-rich repeat kinase 2 exacerbates neuronal cytotoxicity through phosphorylation of histone deacetylase 3 and histone deacetylation. Human Molecular Genetics 2016, 26, 1-18, 10.1093/hmg/ddw363.
- Hyo-Kyoung Choi; Youngsok Choi; Heebum Kang; Eun-Jin Lim; Soo-Yeon Park; Hyun-Seob Lee; Ji-Min Park; Jisook Moon; Yoon-Jung Kim; Insup Choi; et al.Eun-Hye JoeKyung-Chul ChoiKim Yoon-Jung PINK1 positively regulates HDAC3 to suppress dopaminergic neuronal cell death. Human Molecular Genetics 2014, 24, 1127-1141, 10.1093/hmg/ddu526.
- Song, C.; Kanthasamy, A.; Anantharam, V.; Sun, F.; Kanthasamy, A.G. Environmental neurotoxic pesticide increases histone acetylation to promote apoptosis in dopaminergic neuronal cells: Relevance to epigenetic mechanisms of neurodegeneration. Mol. Pharmacol. 2010, 77, 621–632.
- Song, C.; Kanthasamy, A.; Jin, H.; Anantharam, V.; Kanthasamy, A.G. Paraquat induces epigenetic changes by promoting histone acetylation in cell culture models of dopaminergic degeneration. Neurotoxicology 2011, 32, 586–595.
- Daniele Cartelli; Alida Amadeo; Alessandra Maria Calogero; Francesca Vittoria Marialuisa Casagrande; Carmelita De Gregorio; MariaRosa Gioria; Naoko Kuzumaki; Ilaria Costa; Jenny Sassone; Andrea Ciammola; et al.Nobutaka HattoriHideyuki OkanoStefano GoldwurmLaurent RoybonGianni PezzoliGraziella Cappelletti Parkin absence accelerates microtubule aging in dopaminergic neurons. Neurobiology of Aging 2018, 61, 66-74, 10.1016/j.neurobiolaging.2017.09.010.
- Ruoxi Wang; Jieqiong Tan; Tingting Chen; Hailong Han; Runyi Tian; Ya Tan; Yiming Wu; Jingyi Cui; Fang Chen; Jie Li; et al.Lu LvXinjie GuanShuai ShangJia-Hong LuZhuohua Zhang ATP13A2 facilitates HDAC6 recruitment to lysosome to promote autophagosome–lysosome fusion. The Journal of Cell Biology 2018, 218, 267-284, 10.1083/jcb.201804165.
- Wencheng Jian; Xinbing Wei; Lin Chen; Ziying Wang; Yu Sun; ShaoWei Zhu; Haiyan Lou; Shaoqi Yan; Xinbing Li; Junlin Zhou; et al.Bin Zhang Inhibition of HDAC6 increases acetylation of peroxiredoxin1/2 and ameliorates 6-OHDA induced dopaminergic injury. Neuroscience Letters 2017, 658, 114-120, 10.1016/j.neulet.2017.08.029.
- Preeti Singh; Peter S. Hanson; Christopher M. Morris; SIRT1 ameliorates oxidative stress induced neural cell death and is down-regulated in Parkinson’s disease. BMC Neuroscience 2017, 18, 1-13, 10.1186/s12868-017-0364-1.
- Yan-Jie Guo; Su-Yan Dong; Xin-Xin Cui; Ya Feng; Te Liu; Ming Yin; Sheng-Han Kuo; Eng-King Tan; Wen-Juan Zhao; Yun-Cheng Wu; et al. Resveratrol alleviates MPTP-induced motor impairments and pathological changes by autophagic degradation of α-synuclein via SIRT1-deacetylated LC3. Molecular Nutrition & Food Research 2016, 60, 2161-2175, 10.1002/mnfr.201600111.
- Shohei Shinozaki; Kyungho Chang; Michihiro Sakai; Nobuyuki Shimizu; Marina Yamada; Tomokazu Tanaka; Harumasa Nakazawa; Fumito Ichinose; Yoshitsugu Yamada; Akihito Ishigami; et al.Ichinose FumitoYasuyoshi OuchiMarlene E. StarrHiroshi SaitoKentaro ShimokadoJonathan S. StamlerMasao Kaneki Inflammatory stimuli induce inhibitory S-nitrosylation of the deacetylase SIRT1 to increase acetylation and activation of p53 and p65. Science Signaling 2014, 7, ra106-ra106, 10.1126/scisignal.2005375.
- Su-Yan Dong; Yan-Jie Guo; Ya Feng; Xin-Xin Cui; Sheng-Han Kuo; Te Liu; Yun-Cheng Wu; The epigenetic regulation of HIF-1α by SIRT1 in MPP + treated SH-SY5Y cells. Biochemical and Biophysical Research Communications 2016, 470, 453-459, 10.1016/j.bbrc.2016.01.013.
- Feng, Y.; Liu, T.; Dong, S.Y.; Guo, Y.J.; Jankovic, J.; Xu, H.; Wu, Y.C. Rotenone affects p53 transcriptional activity and apoptosis via targeting SIRT1 and H3K9 acetylation in SH-SY5Y cells. J. Neurochem. 2015, 134, 668–676.
- Tao, H.; Liu, Y.; Hou, Y. miRNA3845p regulates the progression of Parkinson’s disease by targeting SIRT1 in mice and SHSY5Y cell. Int. J. Mol. Med. 2020, 45, 441–450.
- Yali Wang; Dongjun Lv; Wenwen Liu; Siyue Li; Jing Chen; Yun Shen; Fen Wang; Li-Fang Hu; Chun-Feng Liu; Disruption of the Circadian Clock Alters Antioxidative Defense via the SIRT1-BMAL1 Pathway in 6-OHDA-Induced Models of Parkinson’s Disease. Oxidative Medicine and Cellular Longevity 2018, 2018, 1-11, 10.1155/2018/4854732.
- Andrew J. Schwab; Samantha L. Sison; Michael R. Meade; Katarzyna A. Broniowska; John A. Corbett; Allison D. Ebert; Decreased Sirtuin Deacetylase Activity in LRRK2 G2019S iPSC-Derived Dopaminergic Neurons. Stem Cell Reports 2017, 9, 1839-1852, 10.1016/j.stemcr.2017.10.010.
- Rita Machado De Oliveira; Hugo Vicente Miranda; Laetitia Francelle; Raquel Pinho; Éva M. Szegö; Renato Martinho; Francesca Munari; Diana F. Lázaro; Sébastien Moniot; Patrícia Guerreiro; et al.Luis Fonseca-OrnelasZrinka MarijanovicPedro AntasEllen GerhardtFrancisco Javier EnguitaBruno FauvetDeborah PenqueTeresa Faria PaisQiang TongStefan BeckerSebastian KüglerHilal Ahmed LashuelClemens SteegbornMarkus ZweckstetterTiago Fleming Outeiro The mechanism of sirtuin 2–mediated exacerbation of alpha-synuclein toxicity in models of Parkinson disease. PLOS Biology 2017, 15, e2000374, 10.1371/journal.pbio.2000374.
- A.R. Esteves; A.M. Palma; R. Gomes; D. Santos; D.F. Silva; S.M. Cardoso; Acetylation as a major determinant to microtubule-dependent autophagy: Relevance to Alzheimer's and Parkinson disease pathology. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2019, 1865, 2008-2023, 10.1016/j.bbadis.2018.11.014.
- Lei Liu; Anirudh Arun; Lakia Ellis; Carina Peritore; Gizem Donmez; SIRT2 enhances 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced nigrostriatal damage via apoptotic pathway. Frontiers in Aging Neuroscience 2014, 6, 184, 10.3389/fnagi.2014.00184.
- Vivek P. Patel; Charleen T. Chu; Decreased SIRT2 activity leads to altered microtubule dynamics in oxidatively-stressed neuronal cells: Implications for Parkinson's disease. Experimental Neurology 2014, 257, 170-181, 10.1016/j.expneurol.2014.04.024.
- Han Shi; Han-Xiang Deng; David Gius; Paul T. Schumacker; D. James Surmeier; Yong-Chao Ma; Sirt3 protects dopaminergic neurons from mitochondrial oxidative stress. Human Molecular Genetics 2017, 26, 1915-1926, 10.1093/hmg/ddx100.
- Xuefei Zhang; Xiaoqing Ren; Qi Zhang; Zheyi Li; Shuaipeng Ma; Jintao Bao; Zeyang Li; Xue Bai; Liangjun Zheng; Zhong Zhang; et al.Shujiang ShangChen ZhangChuangui WangLiu CaoQingsong WangJianguo Ji PGC-1α/ERRα-Sirt3 Pathway Regulates DAergic Neuronal Death by Directly Deacetylating SOD2 and ATP Synthase β. Antioxidants & Redox Signaling 2016, 24, 312-328, 10.1089/ars.2015.6403.
- Xin-Xin Cui; Xuan Li; Su-Yan Dong; Yan-Jie Guo; Te Liu; Yun-Cheng Wu; SIRT3 deacetylated and increased citrate synthase activity in PD model. Biochemical and Biophysical Research Communications 2017, 484, 767-773, 10.1016/j.bbrc.2017.01.163.
- Jae-Hyeon Park; Jeremy D. Burgess; Ayman H. Faroqi; Natasha N. DeMeo; Fabienne C. Fiesel; Wolfdieter Springer; Marion Delenclos; Pamela J. McLean; Alpha-synuclein-induced mitochondrial dysfunction is mediated via a sirtuin 3-dependent pathway. Molecular Neurodegeneration 2020, 15, 1-19, 10.1186/s13024-019-0349-x.




