Acetylation of lysine residues is a key post-translational modification for protein functions in all eukaryotic organisms. Acetylation of lysine residues can be catalyzed by lysine acetyltransferases (KATs) or modified by abundant Ac-CoA through nonenzymatic mechanisms. Conversely, lysine deacetylation is catalyzed by lysine deacetylases (KDACs).
- Lysine Acetylation
- lysine acetyltransferases
- lysine deacetylases
1. Introduction
Acetylation of lysine residues originally discovered in 1964 as a unique post-translational modification of histones, modifications of lysine acetylation and deacetylation are now found in thousands of nonhistone proteins which are located in virtually every cellular compartment and have essential roles in various cellular processes including gene regulation, cell signaling, and metabolism, as well as contribute to the progression of multiple diseases.
2. Lysine Acetylation and Its Regulatory Mechanism and Functions
2.1. Lysine Acetylation and Its Functions
Lysine acetylation in histones was first described by Vincent Allfrey and his colleagues in 1964[1] [15]. Lysine acetylation is an evolutionarily conserved and reversible posttranslational modification (PTM) in eukaryotes that precisely governs protein functions and involves transfer of an acetyl group donated by acetyl coenzyme A (Ac-CoA) to the ε-amino side chain of a protein lysine residue. Lysine acetylation occurs in both histones and nonhistone proteins. Lysine acetylation of histones such as Histone 2A (H2A), Histone 2B (H2B), Histone 3 (H3), and Histone 4 (H4) generally results in transcriptional activation due to destabilization of DNA-histone binding, as acetylation of lysine neutralizes its positive charge, which prevents the formation of salt bridges with the negatively charged phosphate backbone of DNA[2] [16]. In addition to histones, many nonhistone proteins in the cytoplasm and organelles are also dynamically acetylated and deacetylated; these changes are closely implicated in the regulation of various cellular processes, including gene transcription; cell cycle progression; DNA damage repair; cellular signal transduction; protein folding stability and aggregation; cytoskeleton organization; RNA processing and stability; and autophagy regulation[3][4] [17,18].
Acetylation of lysine residues can be catalyzed by KATs or modified by abundant Ac-CoA through nonenzymatic mechanisms. Conversely, lysine deacetylation is catalyzed by KDACs, which comprise two major groups with distinct catalytic mechanisms: NAD+-dependent Sirtuins (SIRTs) and Zn2+-dependent histone deacetylases (HDACs) (Figure 1). The acetylation levels of lysines are highly dynamic, and the balance between lysine acetylation and deacetylation is precisely controlled by KATs and KDACs as well as by the concentration of Ac-CoA in organellar compartments such as mitochondria[5][6] [19,20].
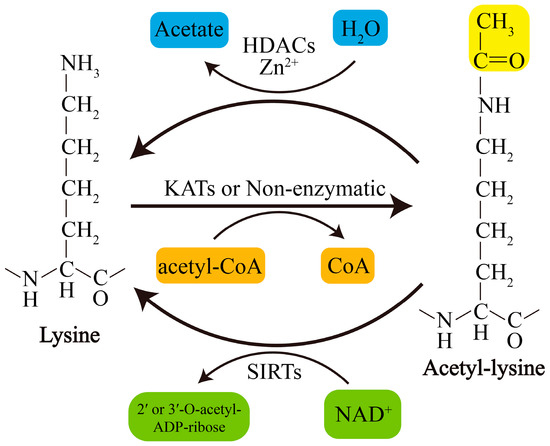
Figure 1. Schematic overview of lysine acetylation and deacetylation. Lysine acetylation, which is catalyzed by KATs, involves transfer of an acetyl group from Ac-CoA to the ε-amino side chain of lysine or occurs nonenzymatically. Deacetylation of lysine residues is catalyzed by Zn2+-dependent HDACs or by NAD+-dependent SIRTs. NAD+, nicotinamide adenine dinucleotide.
2.2. KATs and KDACs in Humans and Their Involvement in PD
To date, at least 22 human KATs have been identified to display acetyltransferase activity; these KATs can be divided into three major families: the MYST family, the GNAT family, and the p300/CBP family[4][7] [18,21] (Table 12). The substrate specificity of KATs is primarily determined by their subcellular distribution or interacting partners or by the availability of lysine in substrates[3] [17]. Most KATs are localized mainly in the nucleus, where they mediate processes including but not limited to histone acetylation, and some KATs also located in the cytoplasm are responsible for cytoplasmic substrate acetylation[3] [17]. Recently, GCN5-like 1 (GCN5L1) and Ac-CoA Acetyltransferase 1 (ACAT1) were identified as mitochondrial KATs that regulate mitochondrial functions by acetylating several mitochondrial substrates[8][9] [22,23]. In addition, the well-known nuclear KAT8/MOF is also found to localize to mitochondria and affect mitochondrial functions[10] [24]. The classifications, subcellular localization, involvement in PD models, and relevant substrates of these KATs are presented in Table 12. However, to date, only a small proportion of KATs have been identified to be related to PD (Table 12).
Table 12. Human KATs and their involvement in PD.
|
Family |
Name |
Subcellular Localization |
PD Model/KAT Change/Substrate Acetylation Changes |
|
GNAT |
KAT1/HAT1 |
Nucleus |
Mn/expression ↓/H3 and H4 ↓ [11].[25]. |
|
GNAT |
KAT2A/GCN5 |
Nucleus |
MPP+/activity ↑/PGC-1α ↑ [12].[26]. |
|
GNAT |
KAT2B/PCAF |
Nucleus |
NA |
|
GNAT |
KAT9/ELP3 |
Nucleus/cytoplasm |
NA |
|
GNAT |
αTAT1/ATAT1 |
Cytoplasm/membrane |
LRRK2 knockout/NA/α-tubulin ↑; LRKK2 R1441C or Y1699C/NA/α-tubulin ↓[13][14]. [27,28]. |
|
p300/CBP |
KAT3A/CBP |
Nucleus/cytoplasm |
Dieldrin/expression ↑/H3 and H4 ↑[15]. [29]. |
|
p300/CBP |
KAT3B/p300 |
Nucleus/cytoplasm |
α-syn/expression and activity ↓/NF-κB-p65 or H3 ↓ [16][17].[30,31]. |
|
MYST |
KAT5/TIP60/PLIP |
Nucleus/cytoplasm |
NA |
|
MYST |
KAT6A/MOZ/MYST3 |
Nucleus |
NA |
|
MYST |
KAT6A/MORF/MYST4 |
Nucleus |
NA |
|
MYST |
KAT7/HBO1/MYST2 |
Nucleus |
NA |
|
MYST |
KAT8/MOF/MYST1 |
Nucleus/mitochondria |
NA |
|
Other |
KAT4/TAF1/TAFII250 |
Nucleus |
NA |
|
Other |
KAT12/TFIIC90 |
Nucleus |
NA |
|
Other |
KAT13A/SRC-1/NCOA1 |
Nucleus |
NA |
|
Other |
KAT13B/SRC-3/NCOA3 |
Nucleus/cytoplasm |
NA |
|
Other |
KAT13C/SRC-2/NCOA2 |
Nucleus |
NA |
|
Other |
KAT13D/CLOCK |
Nucleus/cytoplasm |
NA |
|
Other |
ATF-2/CREB2 |
Nucleus/cytoplasm |
NA |
|
Other |
NAT10 |
Nucleus |
NA |
|
Other |
ACAT1 |
Mitochondria |
NA |
|
Other |
GCN5L1 |
Mitochondria |
NA |
↑, upregulation; ↓, downregulated; NA, not available; PGC-1α, peroxisome proliferator-activated receptor γ coactivator-1α; NF-κB, nuclear factor Kappa-B.
KDACs, originally referred to HDACs, were initially discovered to deacetylate histones in 1995[18] [32]. Later, they were also found to regulate nonhistone protein acetylation and cellular functions[19] [33]. Currently, KDACs are grouped into two major types: NAD+-dependent SIRTs (SIRT1-7) and Zn2+-dependent HDACs (HDAC1-11). They can also be divided into four categories according to phylogeny and sequence similarities: Class I, Class IIa, Class IIb, and Class IV (Table 2). Recently, lymphoid enhancer-binding factor 1 (LEF1) and T cell-specific transcription factor 1 (TCF1) were identified as novel KDACs that are not related to the abovementioned types of KDACs[20] [34]. Zn2+-dependent HDACs are primarily distributed in the nucleus or cytoplasm, although HDAC1 and HDAC7 are also found in mitochondria in some types of cells or under certain conditions[21][22] [35,36]. In contrast, some SIRTs, including SIRT3-5, are restricted to the mitochondria, indicating their unique and crucial roles in mitochondria. However, it should be noted that several KDACs, such as that of SIRT4-6 and some class IIa HDACs, display weak or no deacetylase activity or target other types of acylation[3] [17]. For example, SIRT5 exerts the activity of desuccinylase, demalonylase, and deglutarylase[23] [37]; SIRT4 removes the acyl moieties from lysine residues such as methylglutaryl-, hydroxymethylglutaryl- and 3-methylglutaconyl-lysine[24] [38]; SIRT6 functions to deacetylate long-chain fatty acyl groups rather than protein deacetylation[25] [39]; The classifications, subcellular localization of KDACs, as well as their involvement in PD and the relevant acetylation of substrates are presented in Table 23.
Interestingly, the activity or expression levels of nuclear SIRT1 and mitochondrial SIRT3 are consistently decreased in PD tissues and different PD models. The activity or expression levels of nuclear HDAC2 and HDAC3 are increased in most PD models, but the expression levels of HDAC2 are decreased in tissues of PD patients. Furthermore, the activity or expression levels of two main cytoplasmic KDACs, HDAC6 and SIRT2, are downregulated and upregulated in most PD models, respectively (Table 3). Of note, beyond acetylation, several KATs/KDACs have activity of other acylation modifications including propionyl, butyryl, 2-hydroxyisobutyryl, crotonyl, malonyl, succinyl, or glutaryl modification. For example, p300 has crotonyltransferase activity[26] [40], while KAT2A/GCN5 has both crotonyltransferase and uccinyltransferase activity[27][28] [41,42], whereas HDAC1/2/3/8 and SIRT1-3 possess decrotonylating activity[29][30][31] [43–45]. Whether these changes in KATs or KDACs in PD patients or models also cause variation of other acylation modifications, and the roles of these acylation variations in PD pathology, deserve further research.
Table 23. Human KDACs and their involvement in PD.
|
Class |
Name |
Subcellular Localization |
PD Model/KDAC Change/Substrate Acetylation Level Change |
|
I |
HDAC1 |
Nucleus |
Patient tissues, MPTP or MPP+/expression ↓/H2A, H2B, H3 and H4 ↑[32]. [46]. |
|
I |
HDAC2 |
Nucleus |
Patient tissues, MPTP or MPP+/expression ↓/H2A, H2B, H3 and H4 ↑[32] [46]; MPP+/expression ↑/NA [47]; Midiopathic PP+D fibroblasts/expression ↑/H3 ↓; LRRK2 G2109S PD fibroblasts/expression ↑/NA[33]; idiopathic PD fibroblasts/expression ↑/H3 ↓; LRRK2 G2109S PD fibroblasts/expression ↑/H3 ↓ [34].H3 ↓ [48]. |
|
I |
HDAC3 |
Nucleus |
Idiopathic PD fibroblasts/expression ↑/H3 ↓ [48]; Mn/expression [34]; Mn/expression ↑/H3 and H4 ↓ [11]; LRRK2 or mutation/phosphorylation ↑, nuclear translocation ↑ and activity ↑/H4 ↓ [35]; PINK1 mutation/phosphorylation ↓ and activity ↓/p53 ↑[36].↑/H3 and H4 ↓ [25]; LRRK2 or mutation/phosphorylation ↑, nuclear translocation ↑ and activity ↑/H4 ↓ [49]; PINK1 mutation/phosphorylation ↓ and activity ↓/p53 ↑ [50]. |
|
I |
HDAC8 |
Nucleus/cytoplasm |
NA |
|
IIa |
HDAC4 |
Nucleus/cytoplasm |
Patient tissues, MPTP or MPP+/expression ↓/H2A, H2B, H3 and H4 ↑[32]; [46]; paraquat/expression ↓/H3 ↑ [51,52]; idiopathic PD fibroblasts/expression ↑/H3 ↓, LRRK2 G2109S PD pafibraquatoblasts/expression ↓/H3 ↑[37][38]; idiopathic PD fibroblasts/expression ↑/H3 ↓, LRRK2 G2109S PD fibroblasts/expression ↑/H3 ↓ [48]; Mn/expression ↑/H3 and H4 ↓ [11].↑/H3 ↓ [48]; Mn/expression ↑/H3 and H4 ↓ [25]. |
|
IIa |
HDAC5 |
Nucleus/cytoplasm |
NA |
|
IIa |
HDAC7 |
Nucleus/cytoplasm |
|
|
IIa |
HDAC9 |
Nucleus/cytoplasm |
NA |
|
IIb |
HDAC6 |
Primarily cytoplasm |
Patient tissues, MPTP or MPP+/expression ↓[32]; [46]; idiopathic PD fibroblasts/expression ↓, LRRK2 idiopathicG2109S PD fibroblasts/expression ↓, [48]; Parkin absence/NA/α-tubulin ↑ [53]; ATP13A absence/activity ↓/α-tubulin ↑ [54]; 6-OHDA/expression ↑/peroxiredoxin 1/2 ↓ LRRK2 G2109S PD fibroblasts/expression ↓ [34]; Parkin absence/NA/α-tubulin ↑ [39]; ATP13A absence/activity ↓/α-tubulin ↑[40]; 6-OHDA/expression ↑/peroxiredoxin 1/2 ↓ [41].[55]. |
|
IIb |
HDAC10 |
Primarily cytoplasm |
NA |
|
III |
SIRT1 |
Nucleus |
Patient tissues, MPTP or MPP+/expression ↓/H2A, H2B, H3 and H4 ↑ [46]; [32]patient tissues/activity ↓ [56]; MPTP/expression ↓/LC3 ↑ [57]; MPTP/S-nitrosylation ↑ and activity ↓/p53 and NFκB-p65 ↑ [58]; MPP+/expression ↓/H3 and PGC-1α ↑ [59]; parotient tissues/activity ↓enone/expression ↓/H3 ↑ [60,61]; 6-OHDA/expression ↓/BMAL1 [42];↑ [62]; LRRK2 G2019S MiPTP/expression ↓/LCSC-derived dopaminergic cultures/activity ↓/p53 ↑ [43]; MPTP/S-nitrosylation ↑ and activity ↓/p53 and NFκB-p65 ↑ [44]; MPP+/expression ↓/H3 and PGC-1α ↑[45]; rotenone/expression ↓/H3 ↑[46][47]; 6-OHDA/expression ↓/BMAL1 ↑ [48]; LRRK2 G2019S iPSC-derived dopaminergic cultures/activity ↓/p53 ↑ [49].[63]. |
|
III |
SIRT2 |
Cytoplasm |
MPTP/activity ↑/α-syn ↓[50] [64]; α-syn/activity ↑/α-tubulin ↓ [65]; MPTP or MPP+/activity ↑/Foxo3a ↓ [664]; α-syn]; 6-OHDA/activity ↑↓/α-tubulin ↓[51]; MPTP or MPP+/activity ↑/Foxo3a ↓ [52]; 6-OHDA/activity ↓/α-tubulin ↑[53]. ↑ [67]. |
|
III |
SIRT3 |
Mitochondria |
Patients/NA/MnSOD ↑[54] [68]; MPTP/expression ↓/SOD2 and ATP5B ↑ [69]; MPP+/expression ↓/citrate synthase and isocitrate dehydrogenase 2 ↑ [70]; MPTPα-syn/expression ↓/SOD2 and ATP5B↑ [71]; ↑[55];LRRK2 G2019S MiPP+/expression ↓/citrate synthase and isocitrate dehydrogenase 2 ↑[56]; α-syn/expression ↓/SOD2 ↑ [57]; LRRK2 G2019S iPSC-derived dopaminergic cultures/activity ↓/SOD2 ↑ [49].SC-derived dopaminergic cultures/activity ↓/SOD2 ↑ [63]. |
|
III |
SIRT4 |
Mitochondria |
NA |
|
III |
SIRT5 |
Mitochondria |
NA |
|
III |
SIRT6 |
Nucleus |
NA |
|
III |
SIRT7 |
Nucleolus |
NA |
|
IV |
HDAC11 |
Primarily nucleus |
NA |
|
Other |
TCF1 |
Nucleus |
NA |
|
Other |
LEF1 |
Nucleus |
NA |
↑, upregulation; ↓, downregulated; NA, not available, Mn, manganese; BMAL1, brain and muscle arnt-like 1; iPSC, induced pluripotent stem cells, Foxo3a, Forkhead box O3; ATP5B, ATP synthase subunit β; SOD2, superoxide dismutase.
