
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jeong Hee Hong | -- | 1831 | 2022-06-08 23:57:28 | | | |
| 2 | Peter Tang | Meta information modification | 1831 | 2022-06-09 03:52:11 | | |
Video Upload Options
Nanomaterials serve as carriers for transporting conventional drugs or proteins through lysosomes to various cellular targets. The basic function of lysosomes is to trigger degradation of proteins and lipids. Understanding of lysosomal functions is essential for enhancing the efficacy of nanoparticles-mediated therapy and reducing the malfunctions of cellular metabolism. The lysosomal function is modulated by the movement of ions through various ion channels.
1. Lysosomal Target of Nanoparticles (NPs) and Modulation of NPs for Lysosomal Function
1.1. pH Alteration
1.2. Cell Viability
1.3. Protein Activity and Expression
1.4. Accumulation of NPs
2. Regulation of Lysosomal pH and Its Physiological Function
3. Lysosome-Associated Ion Channels for Lysosomal Function
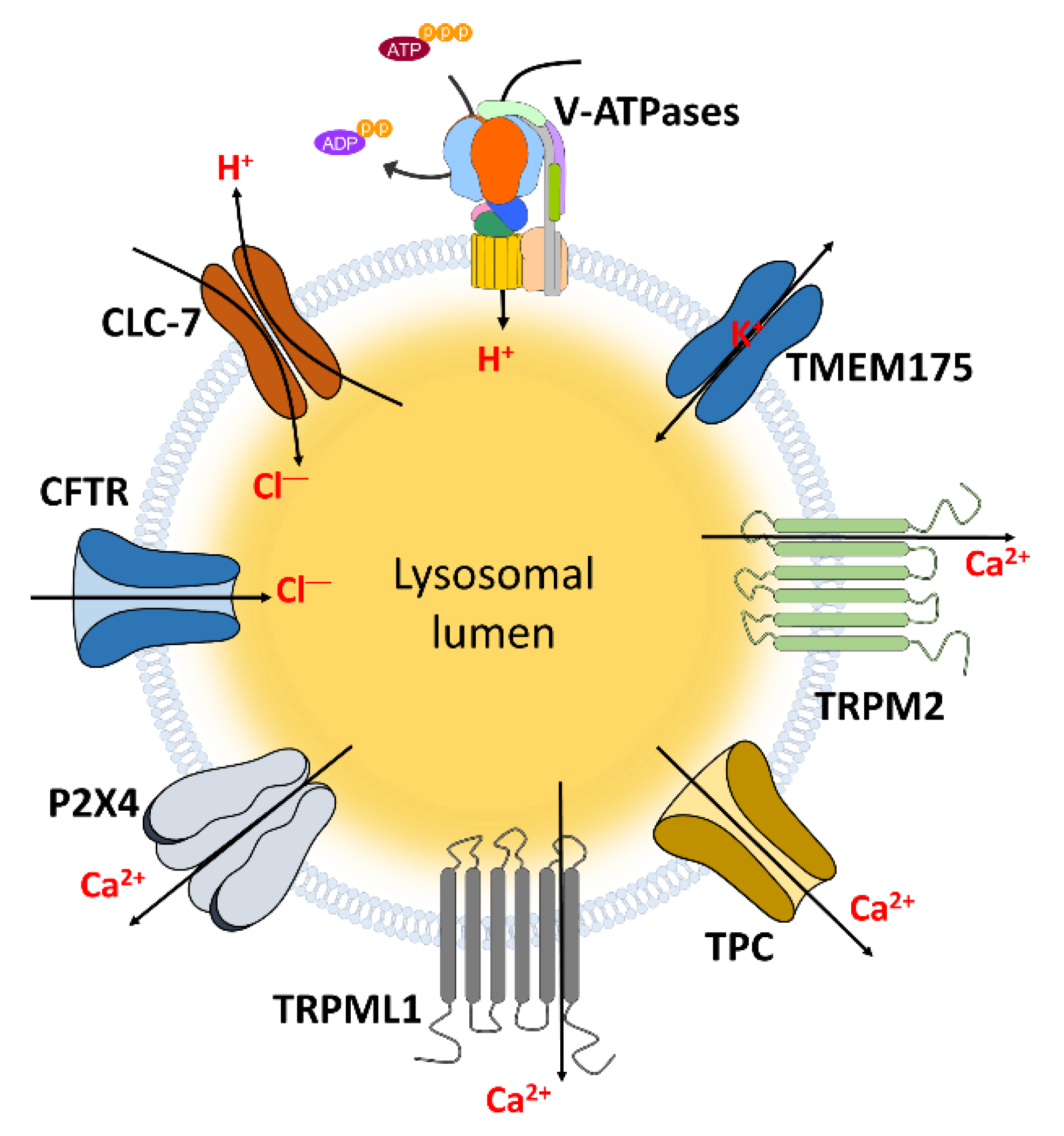
|
Channels |
Mechanisms and Related Diseases |
Ref. |
|---|---|---|
|
CLC-3 |
Promotion of lysosomal acidification |
|
|
CLC-6 |
LSD in CLC-6 mutated neuronal cells |
[67] |
|
CLC-7 |
Maintenance of acidic pH of lysosomes |
|
|
Decrease of dentinogenesis and dental bone formation in CLC-7 deficient mice |
||
|
Degradation of fAβ which drives AD |
||
|
Osteopetrosis in CLC-7 mutation |
||
|
LSD and neurodegeneration in CLC-7-deficient mice |
||
|
CFTR |
Support lysosomal acidification |
[78] |
|
Decrease of bacteria killing function and phago-lysosomal fusion in macrophage |
[79] |
|
|
TRPM2 |
Induce DC maturation and migration |
[80] |
|
Increase of actin remodeling |
[81] |
|
|
Increase of pancreatic β cell apoptosis |
[82] |
|
|
Increase LMP, NLRP3 inflammasome, and mitochondrial fission on the plasma membrane |
||
|
TRPML1 |
Maintenance of acidic pH of lysosomes |
[85] |
|
Increase of large particle phagocytosis, bone remodeling, gastric acid secretion, and myocytes apoptosis |
||
|
Stomach hypertrophy, hypergastrinemia, LSD, mucolipidosis, NPC, and AD in TRPML1 deficiency |
||
|
TMEM175 |
Support lysosomal Ca2+ signaling and pH regulation |
[97] |
|
Related in LSD |
[98] |
|
|
TPC |
Related in autophagy, cancer cell migration, and cellular pigmentation |
|
|
Related in Parkinson’s disease |
||
|
P2X4 |
Promotion of endo-lysosomal fusion |
|
|
Related in liver fibrogenesis |
[106] |
Abbreviations: CLC: Chloride channel; CFTR: Cystic fibrosis transmembrane conductance regulator; TRPM2: Transient receptor potential melastatin 2; TRPML1: Transient receptor potential mucolipin 1; TMEM175: Transmembrane protein 175; TPC: Two pore channel; AD: Alzheimer’s disease; DC: dendritic cell; LMP: Lysosomal membrane permeabilization; NLRP3: NACHT, LRR and PYD domains-containing protein 3; NPC: Niemann-Pick disease type C.
4. NP-Induced Proton Sponge Effect through Ion Channels in the Tumor System
5. Clinical Application and Limitation of Nanomaterials
|
Related Cellular Function |
NPs |
Details |
Reference |
|---|---|---|---|
|
pH alteration (alkalization of lysosome) |
AuNPs |
Increase of oxidative stress, mitochondrial damage, and decrease cell migration/invasion |
[11] |
|
Accumulation of LC3 and block p62 degradation |
[12] |
||
|
AgNPs |
Decrease of TFEB protein expression |
[13] |
|
|
REONPs |
Activation of IL-1β inflammasome |
[14] |
|
|
Cell viability (cell death) |
PNPs |
Decrease of autophagic flux |
[111] |
|
Decrease of cathepsin release |
[112] |
||
|
SiO2 NPs |
Increase of membrane damage and NLRP inflammasome |
||
|
TiO2 NPs |
Increase of membrane damage |
[114] |
|
|
Gd2O3 NPs |
Increase of membrane damage and necrosis |
[115] |
|
|
Protein activity and expression |
AgNPs |
Decrease of lysosomal protease activities |
[35] |
|
REONPs |
Induce lysosomal imbalance by inhibiting mTORC1 pathway |
[37] |
|
|
ZnO NPs |
Increase of macrophage cell death by inhibiting mTORC1 pathway |
[36] |
|
|
Deglycosylation of LAMP-2 |
[38] |
||
|
Carbon nanotube |
Decrease of SNAP |
[44] |
|
|
Accumulation of NPs |
CuO NPs |
Subsequent cellular damage leading to cell death by agglomeration of lysosomes |
|
|
SiO2 NPs, PNPs |
Induce lysosomal swelling leading to apoptosis |
Abbreviations: AuNP: Gold nanoparticle; AgNP: Silver nanoparticle; REONP: rare earth oxide nanoparticle; PNP: polystyrene nanoparticle; ZnO: Zinc oxide; CuO: Copper oxide; TFEB: Transcription factor EB; IL-1β: interleukin-1β; NLRP: NACHT, LRR and PYD domains-containing protein; mTORC1: rapamycin complex 1; SNAP: synaptosomal-associated protein.
References
- De Duve, C.; Wattiaux, R. Functions of lysosomes. Annu. Rev. Physiol. 1966, 28, 435–492.
- de Duve, C. The lysosome turns fifty. Nat. Cell Biol. 2005, 7, 847–849.
- Perera, R.M.; Zoncu, R. The Lysosome as a Regulatory Hub. Annu. Rev. Cell Dev. Biol. 2016, 32, 223–253.
- Pu, J.; Guardia, C.M.; Keren-Kaplan, T.; Bonifacino, J.S. Mechanisms and functions of lysosome positioning. J. Cell Sci. 2016, 129, 4329–4339.
- Mindell, J.A. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 2012, 74, 69–86.
- Walkley, S.U. Pathogenic mechanisms in lysosomal disease: A reappraisal of the role of the lysosome. Acta Paediatr. 2007, 96, 26–32.
- Hipolito, V.E.B.; Ospina-Escobar, E.; Botelho, R.J. Lysosome remodelling and adaptation during phagocyte activation. Cell Microbiol. 2018, 20.
- Herb, M.; Gluschko, A.; Schramm, M. LC3-associated phagocytosis—The highway to hell for phagocytosed microbes. Semin. Cell Dev. Biol. 2019.
- Folts, C.J.; Scott-Hewitt, N.; Proschel, C.; Mayer-Proschel, M.; Noble, M. Lysosomal Re-acidification Prevents Lysosphingolipid-Induced Lysosomal Impairment and Cellular Toxicity. PLoS Biol. 2016, 14, e1002583.
- Stern, S.T.; Adiseshaiah, P.P.; Crist, R.M. Autophagy and lysosomal dysfunction as emerging mechanisms of nanomaterial toxicity. Part. Fibre Toxicol. 2012, 9, 20.
- Woldemichael, T.; Rosania, G.R. The physiological determinants of drug-induced lysosomal stress resistance. PLoS ONE 2017, 12, e0187627.
- Ma, X.; Wu, Y.; Jin, S.; Tian, Y.; Zhang, X.; Zhao, Y.; Yu, L.; Liang, X.J. Gold nanoparticles induce autophagosome accumulation through size-dependent nanoparticle uptake and lysosome impairment. ACS Nano 2011, 5, 8629–8639.
- Miyayama, T.; Fujiki, K.; Matsuoka, M. Silver nanoparticles induce lysosomal-autophagic defects and decreased expression of transcription factor EB in A549 human lung adenocarcinoma cells. Toxicol. Vitr. 2018, 46, 148–154.
- Li, R.; Ji, Z.; Qin, H.; Kang, X.; Sun, B.; Wang, M.; Chang, C.H.; Wang, X.; Zhang, H.; Zou, H.; et al. Interference in autophagosome fusion by rare earth nanoparticles disrupts autophagic flux and regulation of an interleukin-1beta producing inflammasome. ACS Nano 2014, 8, 10280–10292.
- Winchester, B.G. Lysosomal membrane proteins. Eur. J. Paediatr. Neurol. 2001, 5, 11–19.
- Schwake, M.; Schroder, B.; Saftig, P. Lysosomal membrane proteins and their central role in physiology. Traffic 2013, 14, 739–748.
- Yu, S.; Melia, T.J. The coordination of membrane fission and fusion at the end of autophagosome maturation. Curr. Opin. Cell Biol. 2017, 47, 92–98.
- Nascimbeni, A.C.; Codogno, P.; Morel, E. Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J. 2017, 284, 1267–1278.
- Wong, Y.C.; Kim, S.; Peng, W.; Krainc, D. Regulation and Function of Mitochondria-Lysosome Membrane Contact Sites in Cellular Homeostasis. Trends Cell Biol. 2019.
- Helle, S.C.; Kanfer, G.; Kolar, K.; Lang, A.; Michel, A.H.; Kornmann, B. Organization and function of membrane contact sites. Biochim. Biophys. Acta 2013, 1833, 2526–2541.
- Zoncu, R.; Bar-Peled, L.; Efeyan, A.; Wang, S.; Sancak, Y.; Sabatini, D.M. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011, 334, 678–683.
- Johnson, D.E.; Ostrowski, P.; Jaumouille, V.; Grinstein, S. The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 2016, 212, 677–692.
- Li, P.; Gu, M.; Xu, H. Lysosomal Ion Channels as Decoders of Cellular Signals. Trends Biochem. Sci. 2019, 44, 110–124.
- Nishino, I.; Fu, J.; Tanji, K.; Yamada, T.; Shimojo, S.; Koori, T.; Mora, M.; Riggs, J.E.; Oh, S.J.; Koga, Y.; et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 2000, 406, 906–910.
- Kornak, U.; Kasper, D.; Bosl, M.R.; Kaiser, E.; Schweizer, M.; Schulz, A.; Friedrich, W.; Delling, G.; Jentsch, T.J. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell 2001, 104, 205–215.
- Berkovic, S.F.; Dibbens, L.M.; Oshlack, A.; Silver, J.D.; Katerelos, M.; Vears, D.F.; Lullmann-Rauch, R.; Blanz, J.; Zhang, K.W.; Stankovich, J.; et al. Array-based gene discovery with three unrelated subjects shows SCARB2/LIMP-2 deficiency causes myoclonus epilepsy and glomerulosclerosis. Am. J. Hum. Genet. 2008, 82, 673–684.
- Wang, F.; Gomez-Sintes, R.; Boya, P. Lysosomal membrane permeabilization and cell death. Traffic 2018, 19, 918–931.
- Mrschtik, M.; Ryan, K.M. Lysosomal proteins in cell death and autophagy. FEBS J. 2015, 282, 1858–1870.
- Serrano-Puebla, A.; Boya, P. Lysosomal membrane permeabilization in cell death: New evidence and implications for health and disease. Ann. N. Y. Acad. Sci. 2016, 1371, 30–44.
- Micsenyi, M.C.; Sikora, J.; Stephney, G.; Dobrenis, K.; Walkley, S.U. Lysosomal membrane permeability stimulates protein aggregate formation in neurons of a lysosomal disease. J. Neurosci. 2013, 33, 10815–10827.
- Venkatesan, R.; Park, Y.U.; Ji, E.; Yeo, E.J.; Kim, S.Y. Malathion increases apoptotic cell death by inducing lysosomal membrane permeabilization in N2a neuroblastoma cells: A model for neurodegeneration in Alzheimer’s disease. Cell Death Discov. 2017, 3, 17007.
- Korolchuk, V.I.; Saiki, S.; Lichtenberg, M.; Siddiqi, F.H.; Roberts, E.A.; Imarisio, S.; Jahreiss, L.; Sarkar, S.; Futter, M.; Menzies, F.M.; et al. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 2011, 13, 453–460.
- Noda, T.; Ohsumi, Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998, 273, 3963–3966.
- Eskelinen, E.L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Asp. Med. 2006, 27, 495–502.
- Chen, Y.; Wang, M.; Zhang, T.; Du, E.; Liu, Y.; Qi, S.; Xu, Y.; Zhang, Z. Autophagic effects and mechanisms of silver nanoparticles in renal cells under low dose exposure. Ecotoxicol. Environ. Saf. 2018, 166, 71–77.
- Roy, R.; Singh, S.K.; Chauhan, L.K.; Das, M.; Tripathi, A.; Dwivedi, P.D. Zinc oxide nanoparticles induce apoptosis by enhancement of autophagy via PI3K/Akt/mTOR inhibition. Toxicol. Lett. 2014, 227, 29–40.
- Lin, J.; Shi, S.S.; Zhang, J.Q.; Zhang, Y.J.; Zhang, L.; Liu, Y.; Jin, P.P.; Wei, P.F.; Shi, R.H.; Zhou, W.; et al. Giant Cellular Vacuoles Induced by Rare Earth Oxide Nanoparticles are Abnormally Enlarged Endo/Lysosomes and Promote mTOR-Dependent TFEB Nucleus Translocation. Small 2016, 12, 5759–5768.
- Qin, X.; Zhang, J.; Wang, B.; Xu, G.; Zou, Z. LAMP-2 mediates oxidative stress-dependent cell death in Zn(2+)-treated lung epithelium cells. Biochem. Biophys. Res. Commun. 2017, 488, 177–181.
- Cohignac, V.; Landry, M.J.; Ridoux, A.; Pinault, M.; Annangi, B.; Gerdil, A.; Herlin-Boime, N.; Mayne, M.; Haruta, M.; Codogno, P.; et al. Carbon nanotubes, but not spherical nanoparticles, block autophagy by a shape-related targeting of lysosomes in murine macrophages. Autophagy 2018, 14, 1323–1334.
- Hirokawa, N.; Noda, Y. Intracellular transport and kinesin superfamily proteins, KIFs: Structure, function, and dynamics. Physiol. Rev. 2008, 88, 1089–1118.
- Hollenbeck, P.J.; Swanson, J.A. Radial extension of macrophage tubular lysosomes supported by kinesin. Nature 1990, 346, 864–866.
- Paschal, B.M.; Vallee, R.B. Retrograde transport by the microtubule-associated protein MAP 1C. Nature 1987, 330, 181–183.
- Harada, A.; Takei, Y.; Kanai, Y.; Tanaka, Y.; Nonaka, S.; Hirokawa, N. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J. Cell Biol. 1998, 141, 51–59.
- Yuzaki, M. Snapin snaps into the dynein complex for late endosome-lysosome trafficking and autophagy. Neuron 2010, 68, 4–6.
- Chun, Y.; Kim, J. Autophagy: An Essential Degradation Program for Cellular Homeostasis and Life. Cells 2018, 7, 278.
- Settembre, C.; Ballabio, A. Lysosome: Regulator of lipid degradation pathways. Trends Cell Biol. 2014, 24, 743–750.
- Lawrence, R.E.; Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019, 21, 133–142.
- Ferguson, S.M. Neuronal lysosomes. Neurosci. Lett. 2019, 697, 1–9.
- Miyayama, T.; Matsuoka, M. Involvement of lysosomal dysfunction in silver nanoparticle-induced cellular damage in A549 human lung alveolar epithelial cells. J. Occup. Med. Toxicol. 2016, 11, 1.
- Zhang, J.; Zou, Z.; Wang, B.; Xu, G.; Wu, Q.; Zhang, Y.; Yuan, Z.; Yang, X.; Yu, C. Lysosomal deposition of copper oxide nanoparticles triggers HUVEC cells death. Biomaterials 2018, 161, 228–239.
- Schutz, I.; Lopez-Hernandez, T.; Gao, Q.; Puchkov, D.; Jabs, S.; Nordmeyer, D.; Schmudde, M.; Ruhl, E.; Graf, C.M.; Haucke, V. Lysosomal Dysfunction Caused by Cellular Accumulation of Silica Nanoparticles. J. Biol. Chem. 2016, 291, 14170–14184.
- Wang, F.; Bexiga, M.G.; Anguissola, S.; Boya, P.; Simpson, J.C.; Salvati, A.; Dawson, K.A. Time resolved study of cell death mechanisms induced by amine-modified polystyrene nanoparticles. Nanoscale 2013, 5, 10868–10876.
- Ohkuma, S.; Moriyama, Y.; Takano, T. Identification and characterization of a proton pump on lysosomes by fluorescein-isothiocyanate-dextran fluorescence. Proc. Natl. Acad. Sci. USA 1982, 79, 2758–2762.
- Cipriano, D.J.; Wang, Y.; Bond, S.; Hinton, A.; Jefferies, K.C.; Qi, J.; Forgac, M. Structure and regulation of the vacuolar ATPases. Biochim. Biophys. Acta 2008, 1777, 599–604.
- Hirata, T.; Iwamoto-Kihara, A.; Sun-Wada, G.H.; Okajima, T.; Wada, Y.; Futai, M. Subunit rotation of vacuolar-type proton pumping ATPase: Relative rotation of the G and C subunits. J. Biol. Chem. 2003, 278, 23714–23719.
- Yokoyama, K.; Nakano, M.; Imamura, H.; Yoshida, M.; Tamakoshi, M. Rotation of the proteolipid ring in the V-ATPase. J. Biol. Chem. 2003, 278, 24255–24258.
- Ishida, Y.; Nayak, S.; Mindell, J.A.; Grabe, M. A model of lysosomal pH regulation. J. Gen. Physiol. 2013, 141, 705–720.
- Kasper, D.; Planells-Cases, R.; Fuhrmann, J.C.; Scheel, O.; Zeitz, O.; Ruether, K.; Schmitt, A.; Poet, M.; Steinfeld, R.; Schweizer, M.; et al. Loss of the chloride channel ClC-7 leads to lysosomal storage disease and neurodegeneration. EMBO J. 2005, 24, 1079–1091.
- Checchetto, V.; Teardo, E.; Carraretto, L.; Leanza, L.; Szabo, I. Physiology of intracellular potassium channels: A unifying role as mediators of counterion fluxes? Biochim. Biophys. Acta 2016, 1857, 1258–1266.
- Betz, C.; Hall, M.N. Where is mTOR and what is it doing there? J. Cell Biol. 2013, 203, 563–574.
- Sundler, R. Lysosomal and cytosolic pH as regulators of exocytosis in mouse macrophages. Acta Physiol. Scand. 1997, 161, 553–556.
- Tapper, H.; Sundler, R. Role of lysosomal and cytosolic pH in the regulation of macrophage lysosomal enzyme secretion. Biochem. J. 1990, 272, 407–414.
- Camargo, M.J.; Sumpio, B.E.; Maack, T. Renal hydrolysis of absorbed protein: Influence of load and lysosomal pH. Am. J. Physiol. 1984, 247, F656–F664.
- Smith, M.L.; Greene, A.A.; Potashnik, R.; Mendoza, S.A.; Schneider, J.A. Lysosomal cystine transport. Effect of intralysosomal pH and membrane potential. J. Biol. Chem. 1987, 262, 1244–1253.
- Li, X.; Wang, T.; Zhao, Z.; Weinman, S.A. The ClC-3 chloride channel promotes acidification of lysosomes in CHO-K1 and Huh-7 cells. Am. J. Physiol. Cell Physiol. 2002, 282, C1483–C1491.
- Okamoto, F.; Kajiya, H.; Toh, K.; Uchida, S.; Yoshikawa, M.; Sasaki, S.; Kido, M.A.; Tanaka, T.; Okabe, K. Intracellular ClC-3 chloride channels promote bone resorption in vitro through organelle acidification in mouse osteoclasts. Am. J. Physiol. Cell Physiol. 2008, 294, C693–C701.
- Poet, M.; Kornak, U.; Schweizer, M.; Zdebik, A.A.; Scheel, O.; Hoelter, S.; Wurst, W.; Schmitt, A.; Fuhrmann, J.C.; Planells-Cases, R.; et al. Lysosomal storage disease upon disruption of the neuronal chloride transport protein ClC-6. Proc. Natl. Acad. Sci. USA 2006, 103, 13854–13859.
- Henriksen, K.; Gram, J.; Neutzsky-Wulff, A.V.; Jensen, V.K.; Dziegiel, M.H.; Bollerslev, J.; Karsdal, M.A. Characterization of acid flux in osteoclasts from patients harboring a G215R mutation in ClC-7. Biochem. Biophys. Res. Commun. 2009, 378, 804–809.
- Henriksen, K.; Sorensen, M.G.; Jensen, V.K.; Dziegiel, M.H.; Nosjean, O.; Karsdal, M.A. Ion transporters involved in acidification of the resorption lacuna in osteoclasts. Calcif. Tissue Int. 2008, 83, 230–242.
- Wen, X.; Lacruz, R.S.; Paine, M.L. Dental and Cranial Pathologies in Mice Lacking the Cl(-) /H(+) -Exchanger ClC-7. Anat. Rec. 2015, 298, 1502–1508.
- Guo, J.; Bervoets, T.J.; Henriksen, K.; Everts, V.; Bronckers, A.L. Null mutation of chloride channel 7 (Clcn7) impairs dental root formation but does not affect enamel mineralization. Cell Tissue Res. 2016, 363, 361–370.
- Murphy, M.P.; LeVine, H., 3rd. Alzheimer’s disease and the amyloid-beta peptide. J. Alzheimers Dis. 2010, 19, 311–323.
- Majumdar, A.; Capetillo-Zarate, E.; Cruz, D.; Gouras, G.K.; Maxfield, F.R. Degradation of Alzheimer’s amyloid fibrils by microglia requires delivery of ClC-7 to lysosomes. Mol. Biol. Cell 2011, 22, 1664–1676.
- Jentsch, T.J. Chloride and the endosomal-lysosomal pathway: Emerging roles of CLC chloride transporters. J. Physiol. 2007, 578, 633–640.
- Zhao, Q.; Wei, Q.; He, A.; Jia, R.; Xiao, Y. CLC-7: A potential therapeutic target for the treatment of osteoporosis and neurodegeneration. Biochem. Biophys. Res. Commun. 2009, 384, 277–279.
- Weinert, S.; Jabs, S.; Supanchart, C.; Schweizer, M.; Gimber, N.; Richter, M.; Rademann, J.; Stauber, T.; Kornak, U.; Jentsch, T.J. Lysosomal pathology and osteopetrosis upon loss of H+-driven lysosomal Cl- accumulation. Science 2010, 328, 1401–1403.
- Sartelet, A.; Stauber, T.; Coppieters, W.; Ludwig, C.F.; Fasquelle, C.; Druet, T.; Zhang, Z.; Ahariz, N.; Cambisano, N.; Jentsch, T.J.; et al. A missense mutation accelerating the gating of the lysosomal Cl-/H+-exchanger ClC-7/Ostm1 causes osteopetrosis with gingival hamartomas in cattle. Dis. Model. Mech. 2014, 7, 119–128.
- Haggie, P.M.; Verkman, A.S. Unimpaired lysosomal acidification in respiratory epithelial cells in cystic fibrosis. J. Biol. Chem. 2009, 284, 7681–7686.
- Di, A.; Brown, M.E.; Deriy, L.V.; Li, C.; Szeto, F.L.; Chen, Y.; Huang, P.; Tong, J.; Naren, A.P.; Bindokas, V.; et al. CFTR regulates phagosome acidification in macrophages and alters bactericidal activity. Nat. Cell Biol. 2006, 8, 933–944.
- Sumoza-Toledo, A.; Lange, I.; Cortado, H.; Bhagat, H.; Mori, Y.; Fleig, A.; Penner, R.; Partida-Sanchez, S. Dendritic cell maturation and chemotaxis is regulated by TRPM2-mediated lysosomal Ca2+ release. FASEB J. 2011, 25, 3529–3542.
- Li, F.; Abuarab, N.; Sivaprasadarao, A. Reciprocal regulation of actin cytoskeleton remodelling and cell migration by Ca2+ and Zn2+: Role of TRPM2 channels. J. Cell Sci. 2016, 129, 2016–2029.
- Lange, I.; Yamamoto, S.; Partida-Sanchez, S.; Mori, Y.; Fleig, A.; Penner, R. TRPM2 functions as a lysosomal Ca2+-release channel in beta cells. Sci. Signal. 2009, 2, ra23.
- Abuarab, N.; Munsey, T.S.; Jiang, L.H.; Li, J.; Sivaprasadarao, A. High glucose-induced ROS activates TRPM2 to trigger lysosomal membrane permeabilization and Zn(2+)-mediated mitochondrial fission. Sci. Signal. 2017, 10.
- Katsnelson, M.A.; Lozada-Soto, K.M.; Russo, H.M.; Miller, B.A.; Dubyak, G.R. NLRP3 inflammasome signaling is activated by low-level lysosome disruption but inhibited by extensive lysosome disruption: Roles for K+ efflux and Ca2+ influx. Am. J. Physiol. Cell Physiol. 2016, 311, C83–C100.
- Soyombo, A.A.; Tjon-Kon-Sang, S.; Rbaibi, Y.; Bashllari, E.; Bisceglia, J.; Muallem, S.; Kiselyov, K. TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J. Biol. Chem. 2006, 281, 7294–7301.
- Chandra, M.; Zhou, H.; Li, Q.; Muallem, S.; Hofmann, S.L.; Soyombo, A.A. A role for the Ca2+ channel TRPML1 in gastric acid secretion, based on analysis of knockout mice. Gastroenterology 2011, 140, 857–867.
- Erkhembaatar, M.; Gu, D.R.; Lee, S.H.; Yang, Y.M.; Park, S.; Muallem, S.; Shin, D.M.; Kim, M.S. Lysosomal Ca2+ Signaling is Essential for Osteoclastogenesis and Bone Remodeling. J. Bone Min. Res. 2017, 32, 385–396.
- Xu, M.; Li, X.; Walsh, S.W.; Zhang, Y.; Abais, J.M.; Boini, K.M.; Li, P.L. Intracellular two-phase Ca2+ release and apoptosis controlled by TRP-ML1 channel activity in coronary arterial myocytes. Am. J. Physiol. Cell Physiol. 2013, 304, C458–C466.
- Samie, M.; Wang, X.; Zhang, X.; Goschka, A.; Li, X.; Cheng, X.; Gregg, E.; Azar, M.; Zhuo, Y.; Garrity, A.G.; et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev. Cell 2013, 26, 511–524.
- Shen, D.; Wang, X.; Li, X.; Zhang, X.; Yao, Z.; Dibble, S.; Dong, X.P.; Yu, T.; Lieberman, A.P.; Showalter, H.D.; et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat. Commun. 2012, 3, 731.
- Weiss, N. Cross-talk between TRPML1 channel, lipids and lysosomal storage diseases. Commun. Integr. Biol. 2012, 5, 111–113.
- Zeevi, D.A.; Frumkin, A.; Bach, G. TRPML and lysosomal function. Biochim. Biophys. Acta 2007, 1772, 851–858.
- Dong, X.P.; Cheng, X.; Mills, E.; Delling, M.; Wang, F.; Kurz, T.; Xu, H. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature 2008, 455, 992–996.
- Miedel, M.T.; Rbaibi, Y.; Guerriero, C.J.; Colletti, G.; Weixel, K.M.; Weisz, O.A.; Kiselyov, K. Membrane traffic and turnover in TRP-ML1-deficient cells: A revised model for mucolipidosis type IV pathogenesis. J. Exp. Med. 2008, 205, 1477–1490.
- Zhang, F.; Jin, S.; Yi, F.; Li, P.L. TRP-ML1 functions as a lysosomal NAADP-sensitive Ca2+ release channel in coronary arterial myocytes. J. Cell Mol. Med. 2009, 13, 3174–3185.
- Lee, J.H.; McBrayer, M.K.; Wolfe, D.M.; Haslett, L.J.; Kumar, A.; Sato, Y.; Lie, P.P.; Mohan, P.; Coffey, E.E.; Kompella, U.; et al. Presenilin 1 Maintains Lysosomal Ca(2+) Homeostasis via TRPML1 by Regulating vATPase-Mediated Lysosome Acidification. Cell Rep. 2015, 12, 1430–1444.
- Sterea, A.M.; Almasi, S.; El Hiani, Y. The hidden potential of lysosomal ion channels: A new era of oncogenes. Cell Calcium. 2018, 72, 91–103.
- Feng, X.; Zhao, Z.; Li, Q.; Tan, Z. Lysosomal Potassium Channels: Potential Roles in Lysosomal Function and Neurodegenerative Diseases. CNS Neurol. Disord. Drug Targets 2018, 17, 261–266.
- Sun, W.; Yue, J. TPC2 mediates autophagy progression and extracellular vesicle secretion in cancer cells. Exp. Cell Res. 2018, 370, 478–489.
- Grimm, C.; Bartel, K.; Vollmar, A.M.; Biel, M. Endolysosomal Cation Channels and Cancer-A Link with Great Potential. Pharmaceuticals 2018, 11, 4.
- Lin-Moshier, Y.; Keebler, M.V.; Hooper, R.; Boulware, M.J.; Liu, X.; Churamani, D.; Abood, M.E.; Walseth, T.F.; Brailoiu, E.; Patel, S.; et al. The Two-pore channel (TPC) interactome unmasks isoform-specific roles for TPCs in endolysosomal morphology and cell pigmentation. Proc. Natl. Acad. Sci. USA 2014, 111, 13087–13092.
- Hockey, L.N.; Kilpatrick, B.S.; Eden, E.R.; Lin-Moshier, Y.; Brailoiu, G.C.; Brailoiu, E.; Futter, C.E.; Schapira, A.H.; Marchant, J.S.; Patel, S. Dysregulation of lysosomal morphology by pathogenic LRRK2 is corrected by TPC2 inhibition. J. Cell Sci. 2015, 128, 232–238.
- Rivero-Rios, P.; Gomez-Suaga, P.; Fernandez, B.; Madero-Perez, J.; Schwab, A.J.; Ebert, A.D.; Hilfiker, S. Alterations in late endocytic trafficking related to the pathobiology of LRRK2-linked Parkinson’s disease. Biochem Soc. Trans. 2015, 43, 390–395.
- Cao, Q.; Zhong, X.Z.; Zou, Y.; Murrell-Lagnado, R.; Zhu, M.X.; Dong, X.P. Calcium release through P2X4 activates calmodulin to promote endolysosomal membrane fusion. J. Cell Biol. 2015, 209, 879–894.
- Fois, G.; Winkelmann, V.E.; Bareis, L.; Staudenmaier, L.; Hecht, E.; Ziller, C.; Ehinger, K.; Schymeinsky, J.; Kranz, C.; Frick, M. ATP is stored in lamellar bodies to activate vesicular P2X4 in an autocrine fashion upon exocytosis. J. Gen. Physiol. 2018, 150, 277–291.
- Le Guilcher, C.; Garcin, I.; Dellis, O.; Cauchois, F.; Tebbi, A.; Doignon, I.; Guettier, C.; Julien, B.; Tordjmann, T. The P2X4 purinergic receptor regulates hepatic myofibroblast activation during liver fibrogenesis. J. Hepatol. 2018, 69, 644–653.
- Moore, M.N.; Allen, J.I.; McVeigh, A.; Shaw, J. Lysosomal and autophagic reactions as predictive indicators of environmental impact in aquatic animals. Autophagy 2006, 2, 217–220.
- Meng Lin, M.; Kim, H.H.; Kim, H.; Muhammed, M.; Kyung Kim, D. Iron oxide-based nanomagnets in nanomedicine: Fabrication and applications. Nano Rev. 2010, 1.
- Zhu, J.; He, K.; Dai, Z.; Gong, L.; Zhou, T.; Liang, H.; Liu, J. Self-Assembly of Luminescent Gold Nanoparticles with Sensitive pH-Stimulated Structure Transformation and Emission Response toward Lysosome Escape and Intracellular Imaging. Anal. Chem. 2019, 91, 8237–8243.
- Lee, D.U.; Park, J.Y.; Kwon, S.; Park, J.Y.; Kim, Y.H.; Khang, D.; Hong, J.H. Apoptotic lysosomal proton sponge effect in tumor tissue by cationic gold nanorods. Nanoscale 2019, 11, 19980–19993.
- Song, W.; Popp, L.; Yang, J.; Kumar, A.; Gangoli, V.S.; Segatori, L. The autophagic response to polystyrene nanoparticles is mediated by transcription factor EB and depends on surface charge. J. Nanobiotechnol. 2015, 13, 87.
- Wang, F.; Salvati, A.; Boya, P. Lysosome-dependent cell death and deregulated autophagy induced by amine-modified polystyrene nanoparticles. Open Biol. 2018, 8.
- Jessop, F.; Hamilton, R.F., Jr.; Rhoderick, J.F.; Fletcher, P.; Holian, A. Phagolysosome acidification is required for silica and engineered nanoparticle-induced lysosome membrane permeabilization and resultant NLRP3 inflammasome activity. Toxicol. Appl. Pharm. 2017, 318, 58–68.
- Popp, L.; Tran, V.; Patel, R.; Segatori, L. Autophagic response to cellular exposure to titanium dioxide nanoparticles. Acta Biomater. 2018, 79, 354–363.
- Jin, Y.; Chen, S.; Duan, J.; Jia, G.; Zhang, J. Europium-doped Gd2O3 nanotubes cause the necrosis of primary mouse bone marrow stromal cells through lysosome and mitochondrion damage. J. Inorg. Biochem. 2015, 146, 28–36.




