
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Chi-Meng Tzeng | -- | 1901 | 2022-04-21 11:46:18 | | | |
| 2 | Vivi Li | Meta information modification | 1901 | 2022-04-22 05:54:50 | | |
Video Upload Options
Liquid biopsy with circulating tumor DNA (ctDNA) profiling by next-generation sequencing holds great promise to revolutionize clinical oncology. It relies on the basis that ctDNA represents the real-time status of the tumor genome which contains information of genetic alterations. Compared to tissue biopsy, liquid biopsy possesses great advantages such as a less demanding procedure, minimal invasion, ease of frequent sampling, and less sampling bias. Next-generation sequencing (NGS) methods have come to a point that both the cost and performance are suitable for clinical diagnosis. Thus, profiling ctDNA by NGS technologies is becoming more and more popular since it can be applied in the whole process of cancer diagnosis and management.
1. Introduction
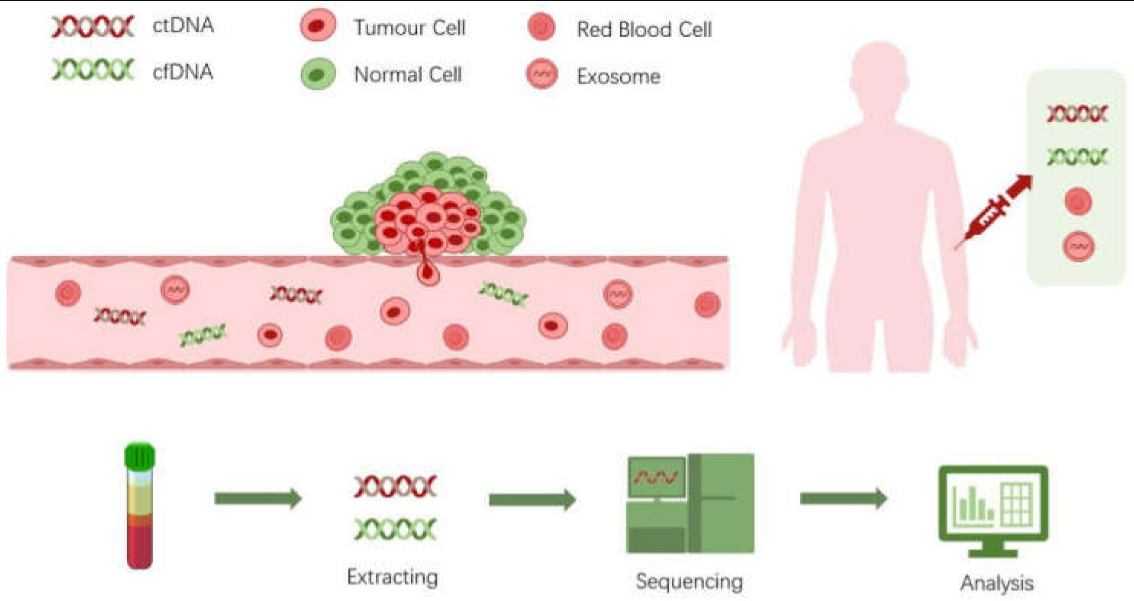
2. Biology of ctDNA
3. Next Generation Sequencing Technologies for ctDNA Detection
4. Strategies for True Mutation Detection Using NGS
References
- Junqueira-Neto, S.; Batista, I.A.; Costa, J.L.; Melo, S.A. Liquid Biopsy beyond Circulating Tumor Cells and Cell-Free DNA. Acta Cytol. 2019, 63, 479–488.
- Poulet, G.; Massias, J.; Taly, V. Liquid Biopsy: General Concepts. Acta Cytol. 2019, 63, 449–455.
- Dakubo, G.D. Cancer Biomarkers in Body Fluids; Springer International Publishing: Cham, Switzerland, 2017.
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351.
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376.
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, eaat4921.
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochim. Biophys. Acta BBA Rev. Cancer 2007, 1775, 181–232.
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.P.S.; Pretorius, P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biol. Rev. 2018, 93, 1649–1683.
- Elazezy, M.; Joosse, S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378.
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841.
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373.
- Avanzini, S.; Kurtz, D.M.; Chabon, J.J.; Moding, E.J.; Hori, S.S.; Gambhir, S.S.; Alizadeh, A.A.; Diehn, M.; Reiter, J.G. A mathematical model of ctDNA shedding predicts tumor detection size. Sci. Adv. 2020, 6, eabc4308.
- Stewart, C.M.; Kothari, P.D.; Mouliere, F.; Mair, R.; Somnay, S.; Benayed, R.; Zehir, A.; Weigelt, B.; Dawson, S.-J.; Arcila, M.E.; et al. The value of cell-free DNA for molecular pathology. J. Pathol. 2018, 244, 616–627.
- Sueoka-Aragane, N.; Nakashima, C.; Yoshida, H.; Matsumoto, N.; Iwanaga, K.; Ebi, N.; Nishiyama, A.; Yatera, K.; Kuyama, S.; Fukuda, M.; et al. The role of comprehensive analysis with circulating tumor DNA in advanced non-small cell lung cancer patients considered for osimertinib treatment. Cancer Med. 2021, 10, 3873–3885.
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437.
- Zaporozhchenko, I.A.; Ponomaryova, A.A.; Rykova, E.Y.; Laktionov, P.P. The potential of circulating cell-free RNA as a can-cer biomarker: Challenges and opportunities. Expert Rev. Mol. Diagn. 2018, 18, 133–145.
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2021, 124, 345–358.
- Lu, L.; Bi, J.; Bao, L. Genetic profiling of cancer with circulating tumor DNA analysis. J. Genet. Genom. 2018, 45, 79–85.
- Kastrisiou, M.; Zarkavelis, G.; Pentheroudakis, G.; Magklara, A. Clinical application of next-generation sequencing as a liq-uid biopsy technique in advanced colorectal cancer: A trick or a treat? Cancers 2019, 11, 1573.
- Bai, Y.; Wang, Z.; Liu, Z.; Liang, G.; Gu, W.; Ge, Q. Technical progress in circulating tumor DNA analysis using next genera-tion sequencing. Mol. Cell Probes 2020, 49, 101480.
- Levy, S.E.; Myers, R.M. Advancements in Next-Generation Sequencing. Annu. Rev. Genom. Hum. Genet. 2016, 17, 95–115.
- Srivastava, V.K. Sequencing Technologies: Introduction and Applications. Int. J. Hum. Genet. 2019, 19.
- Martignano, F.; Munagala, U.; Crucitta, S.; Mingrino, A.; Semeraro, R.; Del Re, M.; Petrini, I.; Magi, A.; Conticello, S.G. Na-nopore sequencing from liquid biopsy: Analysis of copy number variations from cell-free DNA of lung cancer patients. Mol. Cancer 2021, 20, 1–6.
- Chen, S.; Liu, M.; Zhou, Y. Bioinformatics Analysis for Cell-Free Tumor DNA Sequencing Data. Methods Mol. Biol. 2018, 67–95.
- Aparicio-Puerta, E.; Jáspez, D.; Lebrón, R.; Koppers-Lalic, D.; Marchal, J.A.; Hackenberg, M. liqDB: A small-RNAseq knowledge discovery database for liquid biopsy studies. Nucleic Acids Res. 2019, 47, D113–D120.
- Zhao, L.; Wu, X.; Li, T.; Luo, J.; Dong, D. ctcRbase: The gene expression database of circulating tumor cells and microemboli. Database 2020, 2020.
- Cheng, F.; Zhao, J.; Zhao, Z. Advances in computational approaches for prioritizing driver mutations and significantly mutated genes in cancer genomes. Brief. Bioinform. 2016, 17, 642–656.
- Bos, M.K.; Nasserinejad, K.; Jansen, M.P.H.M.; Angus, L.; Atmodimedjo, P.N.; de Jonge, E.; Dinjens, W.N.M.; van Schaik, R.H.N.; Del Re, M.; Dubbink, H.J.; et al. Comparison of variant allele frequency and number of mutant molecules as units of measurement for circulating tumor DNA. Mol. Oncol. 2021, 15, 57–66.
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.Y.; Kaper, F.; Dawson, S.-J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68.
- Gale, D.; Lawson, A.R.J.; Howarth, K.; Madi, M.; Durham, B.; Smalley, S.; Calaway, J.; Blais, S.; Jones, G.; Clark, J.; et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS ONE 2018, 13, e0194630.
- Smith, T.; Heger, A.; Sudbery, I. UMI-tools: Modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017, 27, 491–499.
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535.
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554.
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415.




