
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Daniel S. Dezmirean | -- | 2458 | 2022-04-15 11:48:05 | | | |
| 2 | Peter Tang | Meta information modification | 2458 | 2022-04-18 03:38:03 | | |
Video Upload Options
CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) represents a powerful genome editing technology that revolutionized in a short period of time numerous natural sciences branches. Therefore, extraordinary progress was made in various fields, such as entomology or biotechnology.
1. Introduction
2. The CRISPR-Cas System
2.1. The CRISPR-Cas Complex Role in the Immunity System
2.2. The CRISPR-Cas System as a Genome Editing Tool
|
Traits |
TALEN |
ZFN |
CRISPR-Cas |
References |
|---|---|---|---|---|
|
Origin |
Prokaryotic |
Eukaryotic |
Prokaryotic |
[34] |
|
Efficiency (%) |
76 |
12 |
81 |
[35] |
|
Specificity |
Moderate |
Low |
High |
|
|
Target site recognition |
Any site |
Any site |
Pam motif (NGG) required |
[36] |
|
Multiplex potential |
Low |
Low |
High |
|
|
Processing time |
Time consuming |
Time consuming |
Short |
[38] |
|
Methylation sensitive |
Sensitive |
Sensitive |
Not sensitive |
[35] |
|
Engineering feasibility |
Moderate/High |
Moderate |
Moderate/High |
|
|
Dimerization required |
Yes |
Yes |
No |
[37] |
|
Cost effectiveness |
Moderate |
No |
Yes |
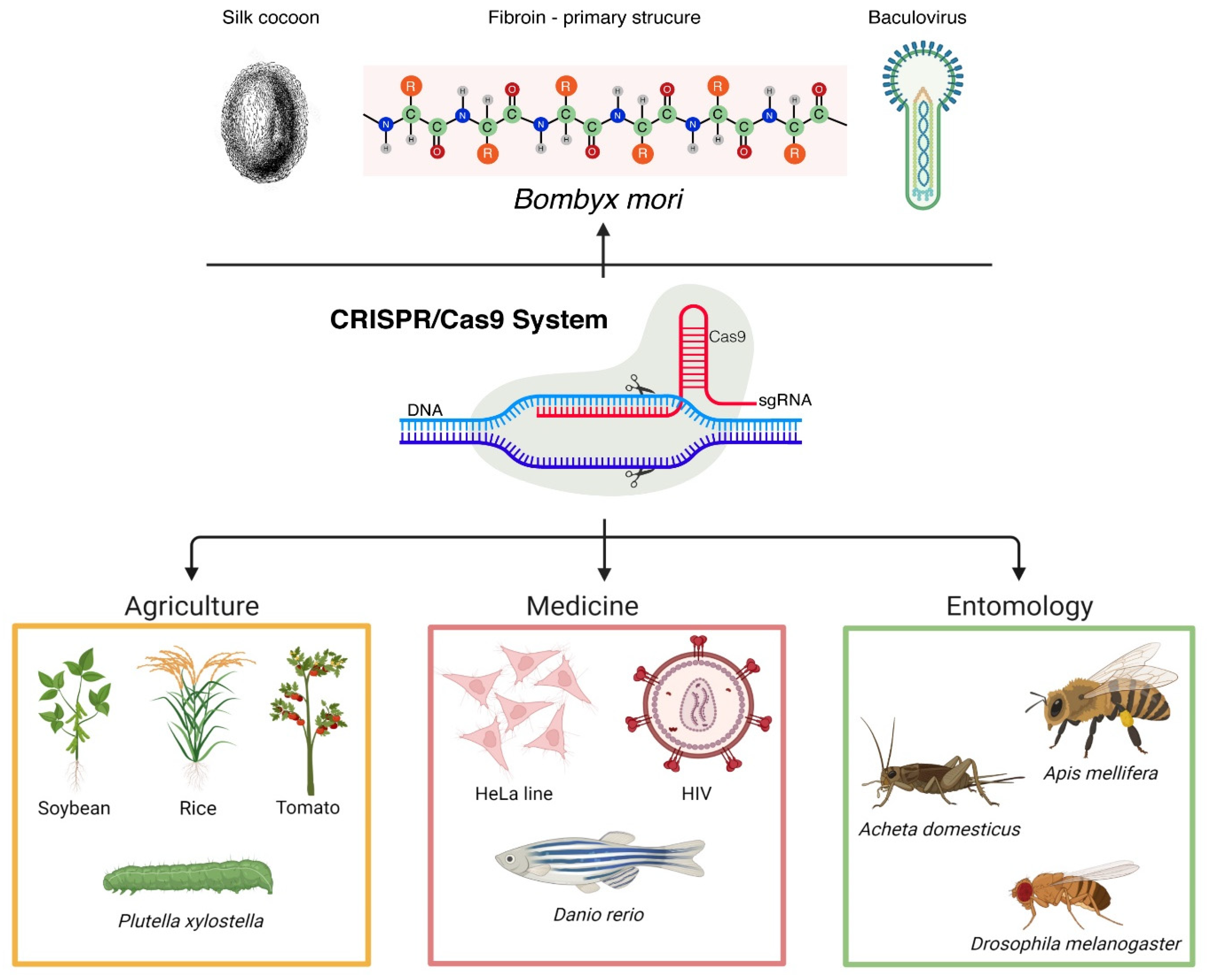
2.3. CRISPR-Cas9 in Entomology
References
- Moon, S.B.; Kim, D.Y.; Ko, J.H.; Kim, Y.S. Recent advances in the CRISPR genome editing tool set. Exp. Mol. Med. 2019, 51, 1–11.
- Guha, T.K.; Wai, A.; Hausner, G. Programmable Genome Editing Tools and their Regulation for Efficient Genome Engineering. Comput. Struct. Biotechnol. J. 2017, 15, 146–160.
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020, 5, 1–23.
- Schuijff, M.; De Jong, M.D.T.; Dijkstra, A.M. AQ methodology study on divergent perspectives on CRISPR-Cas9 in the Netherlands. BMC Med. Ethics 2021, 22, 48.
- Zhang, D.; Hussain, A.; Manghwar, H.; Xie, K.; Xie, S.; Zhao, S.; Larkin, R.M.; Qing, P.; Jin, S.; Ding, F. Genome editing with the CRISPR-Cas system: An art, ethics and global regulatory perspective. Plant Biotechnol. J. 2020, 18, 1651–1669.
- Li, P.; Wang, L.; Yang, J.; Di, L.; Li, J. Applications of the CRISPR-Cas system for infectious disease diagnostics. Expert Rev. Mol. Diagn. 2021, 21, 723–732.
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA—Guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–822.
- Nidhi, S.; Anand, U.; Oleksak, P.; Tripathi, P.; Lal, J.A.; Thomas, G.; Kuca, K.; Tripathi, V. Novel CRISPR—Cas Systems: An Updated Review of the Current Achievements, Applications, and Future Research Perspectives. Int. J. Mol. Sci. 2021, 22, 3327.
- Hahn, F.; Loures, L.S.; Sparks, C.A.; Kanyuka, K.; Nekrasov, V. Efficient CRISPR/Cas-Mediated Targeted Mutagenesis in Spring and Winter Wheat Varieties. Plants 2021, 10, 1481.
- Hesami, M.; Yoosefzadeh Najafabadi, M.; Adamek, K.; Torkamaneh, D.; Jones, A.M.P. Synergizing off-target predictions for in silico insights of CENH3 Knockout in Cannabis through CRISPR/Cas. Molecules 2021, 26, 2053.
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide Sequence of the iap Gene, Responsible for Alkaline Phosphatase Isozyme Conversion in Escherichia coli, and Identification of the Gene Product. J. Bacteriol. 1987, 169, 5429–5433.
- Gophna, U.; Brodt, A. CRISPR/Cas systems in archaea. Mob. Genet. Elem. 2012, 2, 63–64.
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of Bacteria and Archaea. Science 2010, 327, 167–170.
- Shabbir, M.A.B.; Shabbir, M.Z.; Wu, Q.; Mahmood, S.; Sajid, A.; Maan, M.K.; Ahmed, S.; Naveed, U.; Hao, H.; Yuan, Z. CRISPR-cas system: Biological function in microbes and its use to treat antimicrobial resistant pathogens. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 21.
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128.
- Faure, G.; Shmakov, S.A.; Yan, W.X.; Cheng, D.R.; Scott, D.A.; Peters, J.E.; Makarova, K.S.; Koonin, E.V. CRISPR–Cas in mobile genetic elements: Counter-defence and beyond. Nat. Rev. Microbiol. 2019, 17, 513–525.
- Mcdonald, N.D.; Regmi, A.; Morreale, D.P.; Borowski, J.D.; Boyd, E.F. CRISPR-Cas systems are present predominantly on mobile genetic elements in Vibrio species. BMC Genom. 2019, 20, 105.
- Haft, D.H.; Selengut, J.; Mongodin, E.F.; Nelson, K.E. A Guild of 45 CRISPR-Associated (Cas) Protein Families and Multiple CRISPR/Cas Subtypes Exist in Prokaryotic Genomes. PLoS Comput. Biol. 2005, 1, e60.
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477.
- Makarova, K.S.; Koonin, E.V. Annotation and Classification of CRISPR-Cas Systems. In CRISPR; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–27.
- Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. B 2019, 374, 20180087.
- Alkhnbashi, O.S.; Shah, S.A.; Garrett, R.A.; Saunders, S.J.; Costa, F.; Backofen, R. Characterizing leader sequences of CRISPR loci. Bioinformatics 2016, 32, i576–i585.
- Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Garrett, R.A.; Saunders, S.J.; Backofen, R. CRISPRstrand: Predicting repeat orientations to determine the crRNA-encoding strand at CRISPR loci. Bioinformatics 2014, 30, i489–i496.
- McGinn, J.; Marraffini, L.A. Molecular mechanisms of CRISPR–Cas spacer acquisition. Nat. Rev. Microbiol. 2018, 17, 7–12.
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-Mediated Adaptive Immune Systems in Bacteria and Archaea. Annu. Rev. Biochem. 2013, 82, 237–266.
- Roberts, A.; Barrangou, R. Applications of CRISPR-Cas systems in lactic acid bacteria. FEMS Microbio. Rev. 2020, 44, 523–537.
- Hryhorowicz, M.; Lipiński, D.; Zeyland, J.; Słomski, R. CRISPR/Cas9 Immune System as a Tool for Genome Engineering. Arch. Immunol. Ther. Exp. 2016, 65, 233–240.
- Terns, M.P.; Terns, R.M. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011, 14, 321–327.
- Newsom, S.; Parameshwaran, H.P.; Martin, L.; Rajan, R. The CRISPR-Cas Mechanism for Adaptive Immunity and Alternate Bacterial Functions Fuels Diverse Biotechnologies. Front. Cell. Infect. Microbiol. 2021, 10, 1–10.
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010, 11, 181–190.
- Ishino, Y.; Krupovic, M.; Forterre, P. History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. J. Bacteriol. 2018, 200, e00580-17.
- Sontheimer, E.J.; Barrangou, R. The Bacterial Origins of the CRISPR Genome-Editing Revolution. Hum. Gene Ther. 2015, 26, 413–424.
- El-Mounadi, K.; Morales-Floriano, M.L.; Garcia-Ruiz, H. Principles, Applications, and Biosafety of Plant Genome Editing Using CRISPR-Cas9. Front. Plant Sci. 2020, 11, 56.
- Agustın-Pavon, C.; Isalan, M. Synthetic biology and therapeutic strategies for the degenerating brain. Bioessays 2014, 36, 979–990.
- Chen, L.; Tang, L.; Xiang, H.; Jin, L.; Li, Q.; Dong, Y.; Wang, W.; Zhang, G. Advances in genome editing technology and its promising application in evolutionary and ecological studies. Gigascience 2014, 3, 2047-217X.
- Tavakoli, K.; Pour-Aboughadareh, A.; Kianersi, F.; Poczai, P.; Etminan, A.; Shooshtari, L. Applications of CRISPR-Cas9 as an Advanced Genome Editing System in Life Sciences. BioTech 2021, 10, 14.
- Guha, T.K.; Edgell, D.R. Applications of Alternative Nucleases in the Age of CRISPR/Cas9. Int. J. Mol. Sci. 2017, 18, 2565.
- Khan, S.H. Genome-Editing Technologies: Concept, Pros, and Cons of Various Genome-Editing Techniques and Bioethical Concerns for Clinical Application. Mol. Ther. Nucleic Acids 2019, 16, 326–334.
- Chira, S.; Gulei, D.; Hajitou, A.; Zimta, A.A.; Cordelier, P.; Berindan-Neagoe, I. CRISPR/Cas9: Transcending the Reality of Genome Editing. Mol. Ther. Nucleic Acids 2017, 7, 211–222.
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing. Trends Plant Sci. 2019, 24, 1102–1125.
- Li, J.; Shi, Y.; Wu, J.; Li, H.; Smagghe, G.; Liu, T. CRISPR/Cas9 in lepidopteran insects: Progress, application and prospects. J. Insect Physiol. 2021, 135, 104325.
- Tyagi, S.; Kumar, R.; Das, A.; Won, S.Y.; Shukla, P. CRISPR-Cas9 system: A genome-editing tool with endless possibilities. J. Biotechnol. 2020, 319, 36–53.
- Zhang, Y.; Showalter, A.M. CRISPR/Cas9 Genome Editing Technology: A Valuable Tool for Understanding Plant Cell Wall Biosynthesis and Function. Front. Plant Sci. 2020, 11, 589517.
- Collias, D.; Beisel, C.L. CRISPR technologies and the search for the PAM-free nuclease. Nat. Commun. 2021, 12, 1–12.
- Yang, H.; Ren, S.; Yu, S.; Pan, H.; Li, T.; Ge, S.; Zhang, J.; Xia, N. Methods Favoring Homology-Directed Repair Choice in Response to CRISPR/Cas9 Induced-Double Strand Breaks. Int. J. Mol. Sci. 2020, 21, 6461.
- Bernheim, A.; Calvo-villamañán, A.; Basier, C.; Cui, L.; Rocha, E.; Touchon, M.; Bikard, D. Inhibition of NHEJ repair by type II-A CRISPR-Cas systems in bacteria. Nat. Commun. 2017, 8, 25–28.
- Di Stazio, M.; Foschi, N.; Athanasakis, E.; Gasparini, P.; d’Adamo, A.P. Systematic analysis of factors that improve homologous direct repair (HDR) efficiency in CRISPR/Cas9 technique. PLoS ONE 2021, 16, e0247603.
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677.
- Douris, V.; Denecke, S.; Van Leeuwen, T.; Bass, C.; Nauen, R.; Vontas, J. Using CRISPR/Cas9 genome modification to understand the genetic basis of insecticide resistance: Drosophila and beyond. Pestic. Biochem. Physiol. 2020, 167, 104595.
- Tyagi, S.; Kesiraju, K.; Saakre, M.; Rathinam, M.; Raman, V.; Pattanayak, D.; Sreevathsa, R. Genome Editing for Resistance to Insect Pests: An Emerging Tool for Crop Improvement. ACS Omega 2020, 5, 20674–20683.
- Yang, Y.; Xu, J.; Ge, S.; Lai, L. CRISPR/Cas: Advances, Limitations, and Applications for Precision Cancer Research. Front. Med. 2021, 8, 649896.
- Yang, Y.; Liu, X.; Li, S.; Chen, Y.; Zhao, Y.; Wei, Y.; Qiu, Y.; Liu, Y.; Zhou, Z.; Han, J.; et al. Genome-scale CRISPR screening for potential targets of ginsenoside compound K. Cell Death Dis. 2020, 11, 39.
- Xu, Y.; Li, Z. CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy. Comput. Struct. Biotechnol. J. 2020, 18, 2401–2415.
- Ma, X.; He, K.; Shi, Z.; Li, M.; Li, F.; Chen, X.-X. Large-Scale Annotation and Evolution Analysis of MiRNA in Insects. Genome Biol. Evol. 2021, 13, evab083.
- Brady, D.; Grapputo, A.; Romoli, O.; Sandrelli, F. Insect Cecropins, Antimicrobial Peptides with Potential Therapeutic Applications. Int. J. Mol. Sci. 2019, 20, 5862.
- Romoli, O.; Mukherjee, S.; Mohid, S.A.; Dutta, A.; Montali, A.; Franzolin, E.; Brady, D.; Zito, F.; Bergantino, E.; Rampazzo, C.; et al. Enhanced Silkworm Cecropin B Antimicrobial Activity against Pseudomonas aeruginosa from Single Amino Acid Variation. ACS Infect. Dis. 2019, 5, 1200–1213.
- Cui, Y.; Sun, J.; Yu, L. Application of the CRISPR gene-editing technique in insect functional genome studies—A review. Entomol. Exp. Appl. 2017, 162, 124–132.
- De Lazzari, F.; Sandrelli, F.; Whitworth, A.J.; Bisaglia, M. Antioxidant Therapy in Parkinson’s Disease: Insights from Drosophila melanogaster. Antioxidants 2020, 9, 52.
- Taning, C.N.T.; Van Eynde, B.; Yu, N.; Ma, S.; Smagghe, G. CRISPR/Cas9 in insects: Applications, best practices and biosafety concerns. J. Insect Physiol. 2017, 98, 245–257.
- Hu, X.F.; Zhang, B.; Liao, C.H.; Zeng, Z.J. High-Efficiency CRISPR/Cas9-Mediated Gene Editing in Honeybee (Apis mellifera) Embryos. G3 Genes Genomes Genet. 2019, 9, 1759–1766.
- Gratz, S.J.; Cummings, A.M.; Nguyen, J.N.; Hamm, D.C.; Donohue, L.K.; Harrison, M.M.; Wildonger, J.; O’Connor-giles, K.M. Genome Engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 2013, 194, 1029–1035.
- Yu, Z.; Ren, M.; Wang, Z.; Zhang, B.; Rong, Y.S.; Jiao, R.; Gao, G. Highly Efficient Genome Modifications Mediated by CRISPR/Cas9 in Drosophila. Genetics 2013, 195, 289–291.
- Perry, T.; Batterham, P. Harnessing model organisms to study insecticide resistance. Curr. Opin. Insect Sci. 2018, 27, 61–67.
- Homem, R.A.; Davies, T.G.E. An overview of functional genomic tools in deciphering insecticide resistance. Curr. Opin. Insect Sci. 2018, 27, 103–110.
- Scott, J.G.; Buchon, N. Drosophila melanogaster as a powerful tool for studying insect toxicology. Pestic. Biochem. Physiol. 2019, 161, 95–103.
- Matsuoka, Y.; Nakamura, T.; Watanabe, T.; Barnett, A.A.; Noji, S.; Mito, T.; Extavour, C.G. Establishment of CRISPR/Cas9-based knock-in in a hemimetabolous insect: Targeted gene tagging in the cricket Gryllus bimaculatus. bioRxiv 2021.
- Donoughe, S.; Extavour, C.G. Embryonic development of the cricket Gryllus bimaculatus. Dev. Biol. 2016, 411, 140–156.
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656.
- Cucu, A.A.; Baci, G.M.; Moise, A.R.; Dezsi, S.; Marc, B.D.; Stângaciu, S.; Dezmiream, D.S. Towards a Better Understanding of Nutritional and Therapeutic Effects of Honey and Their Applications in Apitherapy. Appl. Sci. 2021, 11, 4190.
- Baci, G.; Cucu, A.; Moise, A.R.; Dezmirean, D.S. Applicability of Honey on Silkworms (Bombyx mori) and Quality Improvement of Its Biomaterials. Appl. Sci. 2021, 11, 4613.
- Kohno, H.; Suenami, S.; Takeuchi, H.; Sasaki, T.; Kubo, T. Production of Knockout Mutants by CRISPR/Cas9 in the European Honeybee, Apis mellifera L. Zool. Sci. 2016, 33, 505–512.
- Nie, H.; Liang, L.; Li, Q.; Li, Z.; Zhu, Y.; Guo, Y.; Zheng, Q.; Lin, Y.; Yang, D.; Li, Z.; et al. CRISPR/Cas9 mediated knockout of Amyellow-y gene results in melanization defect of the cuticle in adult Apis mellifera. J. Insect Physiol. 2021, 132, 104264.
- Gempe, T.; Hasselmann, M.; Schiøtt, M.; Hause, G.; Otte, M.; Beye, M. Sex Determination in Honeybees: Two Separate Mechanisms Induce and Maintain the Female Pathway. PLoS Biol. 2009, 7, e1000222.
- Wang, X.; Lin, Y.; Liang, L.; Geng, H.; Zhang, M.; Nie, H.; Su, S. Transcriptional Profiles of Diploid Mutant Apis mellifera Embryos after Knockout of csd by CRISPR/Cas9. Insects 2021, 12, 704.





