
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Carsten Carlberg | -- | 1772 | 2022-04-13 08:39:43 | | | |
| 2 | Catherine Yang | -2 word(s) | 1770 | 2022-04-13 08:43:27 | | | | |
| 3 | Catherine Yang | Meta information modification | 1770 | 2022-04-15 07:49:30 | | |
Video Upload Options
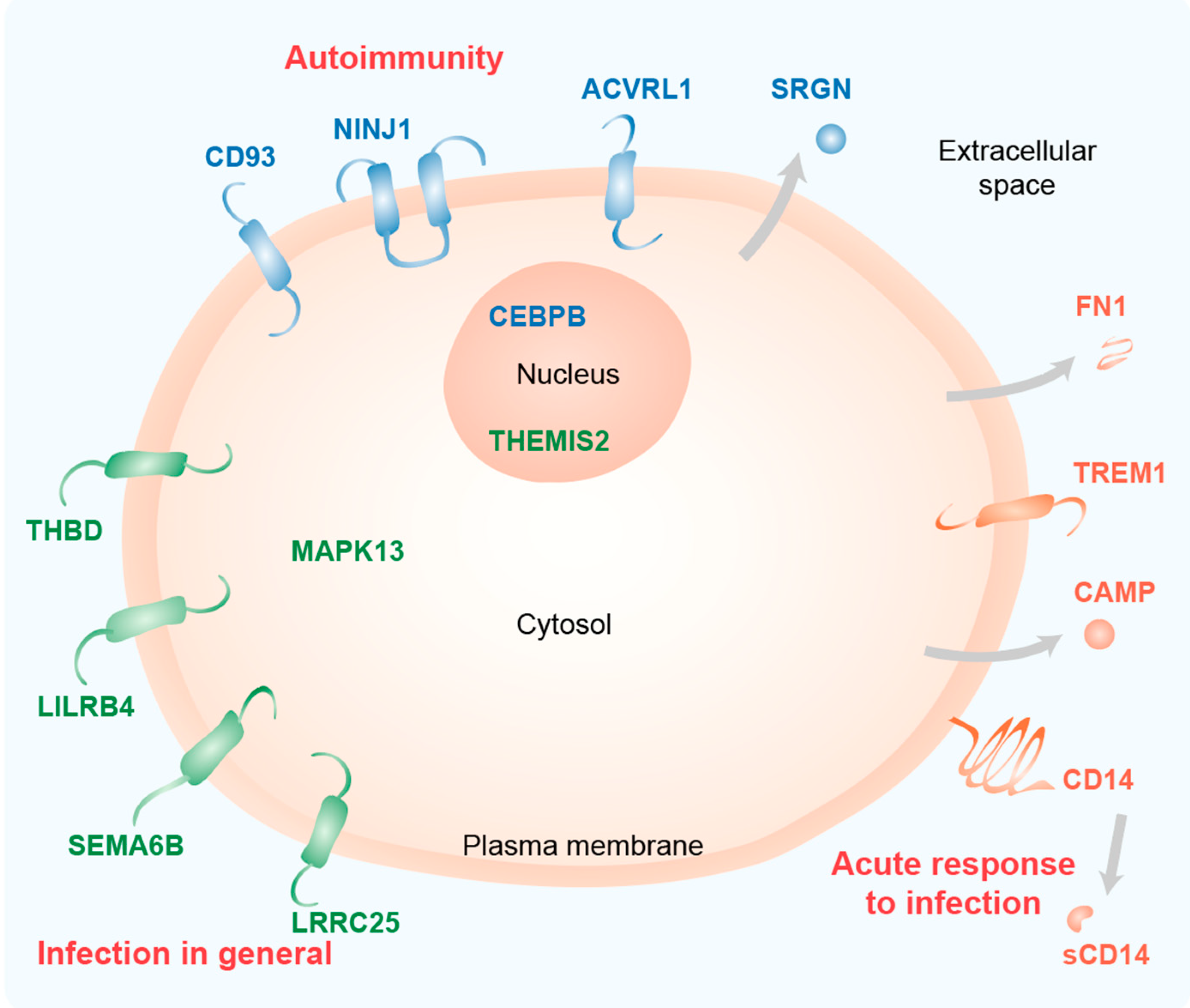
The biologically active form of vitamin D3, 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3), modulates innate and adaptive immunity via genes regulated by the transcription factor vitamin D receptor (VDR). In order to identify the key vitamin D target genes involved in these processes, transcriptome-wide datasets were compared, which were obtained from a human monocytic cell line (THP-1) and peripheral blood mononuclear cells (PBMCs) treated in vitro by 1,25(OH)2D3, filtered using different approaches, as well as from PBMCs of individuals supplemented with a vitamin D3 bolus. The led to the genes ACVRL1, CAMP, CD14, CD93, CEBPB, FN1, MAPK13, NINJ1, LILRB4, LRRC25, SEMA6B, SRGN, THBD, THEMIS2 and TREM1. Public epigenome- and transcriptome-wide data from THP-1 cells were used to characterize these genes based on the level of their VDR-driven enhancers as well as the level of the dynamics of their mRNA production.
1. Introduction
2. Key Vitamin D Target Genes with Functions in the Immune System
References
- Van de Peppel, J.; van Leeuwen, J.P. Vitamin D and gene networks in human osteoblasts. Front. Physiol. 2014, 5, 137.
- Carmeliet, G.; Dermauw, V.; Bouillon, R. Vitamin D signaling in calcium and bone homeostasis: A delicate balance. Best Pract. Res. Clin. Endocrinol. Metab. 2015, 29, 621–631.
- Holick, M.F. Photobiology of Vitamin D. Vitamin D 2018, 45–55.
- Carlberg, C. Molecular endocrinology of vitamin D on the epigenome level. Mol. Cell Endocrinol. 2017, 453, 14–21.
- Carlberg, C. Genome-wide (over)view on the actions of vitamin D. Front. Physiol. 2014, 5, 167.
- Whitfield, G.K.; Dang, H.T.; Schluter, S.F.; Bernstein, R.M.; Bunag, T.; Manzon, L.A.; Hsieh, G.; Dominguez, C.E.; Youson, J.H.; Haussler, M.R.; et al. Cloning of a functional vitamin D receptor from the lamprey (Petromyzon marinus), an ancient vertebrate lacking a calcified skeleton and teeth. Endocrinology 2003, 144, 2704–2716.
- Bouillon, R.; Suda, T. Vitamin D: Calcium and bone homeostasis during evolution. BoneKEy Reports 2014, 3, 480.
- Tsukasaki, M.; Takayanagi, H. Osteoimmunology: Evolving concepts in bone-immune interactions in health and disease. Nat. Rev. Immunol. 2019, 19, 626–642.
- Hanel, A.; Carlberg, C. Vitamin D and evolution: Pharmacologic implications. Biochem. Pharmacol. 2020, 173, 113595.
- Chun, R.F.; Liu, P.T.; Modlin, R.L.; Adams, J.S.; Hewison, M. Impact of vitamin D on immune function: Lessons learned from genome-wide analysis. Front. Physiol. 2014, 5, 151.
- Vintilescu, B.S.; Niculescu, C.E.; Stepan, M.D.; Ionita, E. Involvement of vitamin D in chronic infections of the Waldeyer‘s ring in the school aged child. Curr. Health Sci. J. 2019, 45, 291–295.
- Lu, M.; McComish, B.J.; Burdon, K.P.; Taylor, B.V.; Körner, H. The association between vitamin D and multiple sclerosis risk: 1,25(OH)2D3 induces super-enhancers bound by VDR. Front. Immunol. 2019, 10, 488.
- Caraba, A.; Crisan, V.; Romosan, I.; Mozos, I.; Murariu, M. Vitamin D status, disease activity, and endothelial dysfunction in early rheumatoid arthritis patients. Dis. Markers 2017, 2017, 5241012.
- Holick, M.F. Resurrection of vitamin D deficiency and rickets. J. Clin. Investig. 2006, 116, 2062–2072.
- Grant, W.B.; Lahore, H.; McDonnell, S.L.; Baggerly, C.A.; French, C.B.; Aliano, J.L.; Bhattoa, H.P. Evidence that vitamin D supplementation could reduce risk of influenza and COVID-19 infections and deaths. Nutrients 2020, 12, 988.
- Carlberg, C. Vitamin D signaling in the context of innate immunity: Focus on human monocytes. Front. Immunol. 2019, 10, 2211.
- Carlberg, C. Vitamin D genomics: From in vitro to in vivo. Front. Endocrinol. 2018, 9, 250.
- Carlberg, C.; Molnár, F. Mechanisms of gene regulation. Springer Textbook 2016.
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319.
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637.
- Rando, O.J.; Chang, H.Y. Genome-wide views of chromatin structure. Annu. Rev. Biochem. 2009, 78, 245–271.
- Rivera, C.M.; Ren, B. Mapping human epigenomes. Cell 2013, 155, 39–55.
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936.
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006, 125, 315–326.
- Park, P.J. ChIP-seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009, 10, 669–680.
- Giresi, P.G.; Kim, J.; McDaniell, R.M.; Iyer, V.R.; Lieb, J.D. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome. Res. 2007, 17, 877–885.
- Tuoresmäki, P.; Väisänen, S.; Neme, A.; Heikkinen, S.; Carlberg, C. Patterns of genome-wide VDR locations. PLoS ONE 2014, 9, e96105.
- Neme, A.; Seuter, S.; Carlberg, C. Selective regulation of biological processes by vitamin D based on the spatio-temporal cistrome of its receptor. Biochim. Biophys. Acta 2017, 1860, 952–961.
- Gombart, A.F.; Borregaard, N.; Koeffler, H.P. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. FASEB J. 2005, 19, 1067–1077.
- Liu, P.T.; Stenger, S.; Li, H.; Wenzel, L.; Tan, B.H.; Krutzik, S.R.; Ochoa, M.T.; Schauber, J.; Wu, K.; Meinken, C.; et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 2006, 311, 1770–1773.
- Zanoni, I.; Granucci, F. Role of CD14 in host protection against infections and in metabolism regulation. Front. Cell. Infect. Microbiol. 2013, 3, 32.
- Zanoni, I.; Tan, Y.; Di Gioia, M.; Springstead, J.R.; Kagan, J.C. By capturing inflammatory lipids released from dying cells, the receptor CD14 induces inflammasome-dependent phagocyte hyperactivation. Immunity 2017, 47, 697–709 e693.
- Dower, K.; Ellis, D.K.; Saraf, K.; Jelinsky, S.A.; Lin, L.L. Innate immune responses to TREM-1 activation: Overlap, divergence, and positive and negative cross-talk with bacterial lipopolysaccharide. J. Immunol. 2008, 180, 3520–3534.
- Kim, T.H.; Lee, B.; Kwon, E.; Choi, S.J.; Lee, Y.H.; Song, G.G.; Sohn, J.; Ji, J.D. Regulation of TREM-1 expression by 1,25-dihydroxyvitamin D3 in human monocytes/macrophages. Immunol. Lett. 2013, 154, 80–85.
- Polly, P.; Carlberg, C.; Eisman, J.A.; Morrison, N.A. Identification of a vitamin D3 response element in the fibronectin gene that is bound by a vitamin D3 receptor homodimer. J. Cell Biochem. 1996, 60, 322–333.
- Okamura, Y.; Watari, M.; Jerud, E.S.; Young, D.W.; Ishizaka, S.T.; Rose, J.; Chow, J.C.; Strauss, J.F., 3rd. The extra domain A of fibronectin activates Toll-like receptor 4. J. Biol. Chem. 2001, 276, 10229–10233.
- Loghmani, H.; Conway, E.M. Exploring traditional and nontraditional roles for thrombomodulin. Blood 2018, 132, 148–158.
- Ma, C.Y.; Shi, G.Y.; Shi, C.S.; Kao, Y.C.; Lin, S.W.; Wu, H.L. Monocytic thrombomodulin triggers LPS- and gram-negative bacteria-induced inflammatory response. J. Immunol. 2012, 188, 6328–6337.
- Heikkinen, S.; Väisänen, S.; Pehkonen, P.; Seuter, S.; Benes, V.; Carlberg, C. Nuclear hormone 1α,25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res. 2011, 39, 9181–9193.
- Carlberg, C.; Seuter, S.; de Mello, V.D.; Schwab, U.; Voutilainen, S.; Pulkki, K.; Nurmi, T.; Virtanen, J.; Tuomainen, T.P.; Uusitupa, M. Primary vitamin D target genes allow a categorization of possible benefits of vitamin D3 supplementation. PLoS ONE 2013, 8, e71042.
- Park, M.; Liu, R.W.; An, H.; Geczy, C.L.; Thomas, P.S.; Tedla, N. A dual positive and negative regulation of monocyte activation by leukocyte Ig-like receptor B4 depends on the position of the tyrosine residues in its ITIMs. Innate. Immun. 2017, 23, 381–391.
- Chang, C.C.; Zhang, Q.Y.; Liu, Z.; Clynes, R.A.; Suciu-Foca, N.; Vlad, G. Downregulation of inflammatory microRNAs by Ig-like transcript 3 is essential for the differentiation of human CD8(+) T suppressor cells. J. Immunol. 2012, 188, 3042–3052.
- Waschbisch, A.; Sanderson, N.; Krumbholz, M.; Vlad, G.; Theil, D.; Schwab, S.; Maurer, M.; Derfuss, T. Interferon beta and vitamin D synergize to induce immunoregulatory receptors on peripheral blood monocytes of multiple sclerosis patients. PLoS ONE 2014, 9, e115488.
- Alto, L.T.; Terman, J.R. Semaphorins and their signaling mechanisms. Methods Mol. Biol. 2017, 1493, 1–25.
- Sutton, A.L.; Zhang, X.; Dowd, D.R.; Kharode, Y.P.; Komm, B.S.; Macdonald, P.N. Semaphorin 3B is a 1,25-Dihydroxyvitamin D3-induced gene in osteoblasts that promotes osteoclastogenesis and induces osteopenia in mice. Mol. Endocrinol. 2008, 22, 1370–1381.
- Feng, Y.; Duan, T.; Du, Y.; Jin, S.; Wang, M.; Cui, J.; Wang, R.F. LRRC25 functions as an inhibitor of NF-kappaB signaling pathway by promoting p65/RelA for autophagic degradation. Sci. Rep. 2017, 7, 13448.
- Du, Y.; Duan, T.; Feng, Y.; Liu, Q.; Lin, M.; Cui, J.; Wang, R.F. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J. 2018, 37, 351–366.
- Teymoori-Rad, M.; Shokri, F.; Salimi, V.; Marashi, S.M. The interplay between vitamin D and viral infections. Rev. Med. Virol. 2019, 29, e2032.
- Lee, C. Controversial effects of vitamin D and related genes on viral infections, pathogenesis, and treatment outcomes. Nutrients 2020, 12, 962.
- Mandlik, R.; Mughal, Z.; Khadilkar, A.; Chiplonkar, S.; Ekbote, V.; Kajale, N.; Patwardhan, V.; Padidela, R.; Khadilkar, V. Occurrence of infections in schoolchildren subsequent to supplementation with vitamin D-calcium or zinc: A randomized, double-blind, placebo-controlled trial. Nutr. Res. Pract. 2020, 14, 117–126.
- Urashima, M.; Segawa, T.; Okazaki, M.; Kurihara, M.; Wada, Y.; Ida, H. Randomized trial of vitamin D supplementation to prevent seasonal influenza A in schoolchildren. Am. J. Clin. Nutr. 2010, 91, 1255–1260.
- Risco, A.; del Fresno, C.; Mambol, A.; Alsina-Beauchamp, D.; MacKenzie, K.F.; Yang, H.T.; Barber, D.F.; Morcelle, C.; Arthur, J.S.; Ley, S.C.; et al. p38gamma and p38delta kinases regulate the Toll-like receptor 4 (TLR4)-induced cytokine production by controlling ERK1/2 protein kinase pathway activation. Proc. Natl. Acad. Sci. USA 2012, 109, 11200–11205.
- Ryan, Z.C.; Craig, T.A.; Folmes, C.D.; Wang, X.; Lanza, I.R.; Schaible, N.S.; Salisbury, J.L.; Nair, K.S.; Terzic, A.; Sieck, G.C.; et al. 1α,25-Dihydroxyvitamin D3 regulates mitochondrial oxygen consumption and dynamics in human skeletal muscle cells. J. Biol. Chem. 2016, 291, 1514–1528.
- Lesourne, R.; Zvezdova, E.; Song, K.D.; El-Khoury, D.; Uehara, S.; Barr, V.A.; Samelson, L.E.; Love, P.E. Interchangeability of Themis1 and Themis2 in thymocyte development reveals two related proteins with conserved molecular function. J. Immunol. 2012, 189, 1154–1161.
- Treeck, O.; Odani, T.; Itoh, N.; Imai, H.; Fujita, S.; Kohroki, J.; Nakanishi, T.; Diedrich, K.; Ortmann, O.; Tanaka, K.; et al. Detection of increased icb-1 transcript levels in maturing HL-60 cells: A novel marker for granulocytic and monocytic in vitro differentiation. Leuk Res. 2002, 26, 765–769.
- Duvetorp, A.; Slind Olsen, R.; Skarstedt, M.; Soderman, J.; Seifert, O. Psoriasis and pro-angiogenetic factor CD93: Gene expression and association with gene polymorphism suggests a role in disease pathogenesis. Acta Derm. Venereol. 2017, 97, 916–921.
- Nativel, B.; Ramin-Mangata, S.; Mevizou, R.; Figuester, A.; Andries, J.; Iwema, T.; Ikewaki, N.; Gasque, P.; Viranaicken, W. CD93 is a cell surface lectin receptor involved in the control of the inflammatory response stimulated by exogenous DNA. Immunology 2019, 158, 85–93.
- Griffiths, M.R.; Botto, M.; Morgan, B.P.; Neal, J.W.; Gasque, P. CD93 regulates central nervous system inflammation in two mouse models of autoimmune encephalomyelitis. Immunology 2018, 155, 346–355.
- Jennewein, C.; Sowa, R.; Faber, A.C.; Dildey, M.; von Knethen, A.; Meybohm, P.; Scheller, B.; Drose, S.; Zacharowski, K. Contribution of Ninjurin1 to Toll-like receptor 4 signaling and systemic inflammation. Am. J. Respir. Cell Mol. Biol. 2015, 53, 656–663.
- Ahn, B.J.; Le, H.; Shin, M.W.; Bae, S.J.; Lee, E.J.; Lee, S.Y.; Yang, J.H.; Wee, H.J.; Cha, J.H.; Seo, J.H.; et al. Ninjurin1 enhances the basal motility and transendothelial migration of immune cells by inducing protrusive membrane dynamics. J. Biol. Chem. 2014, 289, 21926–21936.
- Simpson-Abelson, M.R.; Hernandez-Mir, G.; Childs, E.E.; Cruz, J.A.; Poholek, A.C.; Chattopadhyay, A.; Gaffen, S.L.; McGeachy, M.J. CCAAT/Enhancer-binding protein beta promotes pathogenesis of EAE. Cytokine 2017, 92, 24–32.
- Marchwicka, A.; Marcinkowska, E. Regulation of expression of CEBP genes by variably expressed vitamin D receptor and retinoic acid receptor alpha in human acute myeloid leukemia cell lines. Int. J. Mol. Sci. 2018, 19, 1918.
- Bocci, M.; Sjolund, J.; Kurzejamska, E.; Lindgren, D.; Marzouka, N.A.; Bartoschek, M.; Hoglund, M.; Pietras, K. Activin receptor-like kinase 1 is associated with immune cell infiltration and regulates CLEC14A transcription in cancer. Angiogenesis 2019, 22, 117–131.
- Ojeda-Fernandez, L.; Recio-Poveda, L.; Aristorena, M.; Lastres, P.; Blanco, F.J.; Sanz-Rodriguez, F.; Gallardo-Vara, E.; de las Casas-Engel, M.; Corbi, A.; Arthur, H.M.; et al. Mice lacking endoglin in macrophages show an impaired immune response. PLoS Genet 2016, 12, e1005935.
- Korpetinou, A.; Skandalis, S.S.; Labropoulou, V.T.; Smirlaki, G.; Noulas, A.; Karamanos, N.K.; Theocharis, A.D. Serglycin: At the crossroad of inflammation and malignancy. Front. Oncol. 2014, 3, 327.
- Kolseth, I.B.; Reine, T.M.; Vuong, T.T.; Meen, A.J.; Fan, Q.; Jenssen, T.G.; Gronning-Wang, L.M.; Kolset, S.O. Serglycin is part of the secretory repertoire of LPS-activated monocytes. Immun. Inflamm. Dis. 2015, 3, 23–31.




