Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Néstor Carrillo | -- | 1851 | 2022-04-01 16:05:25 | | | |
| 2 | Camila Xu | Meta information modification | 1851 | 2022-04-02 04:00:20 | | | | |
| 3 | Camila Xu | -2 word(s) | 1849 | 2022-04-02 04:03:14 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Carrillo, N.; Mayta, M.L.; Hajirezaei, M.; Lodeyro, A.F. Leaf Senescence. Encyclopedia. Available online: https://encyclopedia.pub/entry/21293 (accessed on 07 February 2026).
Carrillo N, Mayta ML, Hajirezaei M, Lodeyro AF. Leaf Senescence. Encyclopedia. Available at: https://encyclopedia.pub/entry/21293. Accessed February 07, 2026.
Carrillo, Néstor, Martín Leonardo Mayta, Mohammad-Reza Hajirezaei, Anabella Fernanda Lodeyro. "Leaf Senescence" Encyclopedia, https://encyclopedia.pub/entry/21293 (accessed February 07, 2026).
Carrillo, N., Mayta, M.L., Hajirezaei, M., & Lodeyro, A.F. (2022, April 01). Leaf Senescence. In Encyclopedia. https://encyclopedia.pub/entry/21293
Carrillo, Néstor, et al. "Leaf Senescence." Encyclopedia. Web. 01 April, 2022.
Copy Citation
Leaf senescence is an ordered physiological process in which cellular structures and biomolecules are progressively broken down and the resulting products mobilized to other plant organs such as fruits, seeds, tubers and/or more apical leaves.
senescence
reactive oxygen species
chloroplast
phytohormones
1. Introduction
Leaf senescence is an ordered physiological process in which cellular structures and biomolecules are progressively broken down and the resulting products mobilized to other plant organs such as fruits, seeds, tubers and/or more apical leaves [1][2][3]. The most visible manifestation of leaf senescence is yellowing caused by destruction of the chloroplast pigment-protein complexes and conversion of the constituent chlorophylls (Chl) into catabolic non-green derivatives after opening of the chlorin ring system [4][5]. Natural leaf senescence as it occurs in the field is normally age-dependent and accelerates upon transition of the vegetative into the reproductive growth phase [6][7].
Senescence is controlled by a genetic program involving major changes in expression patterns that result in degradation of cells targeted for demise and reallocation of the resulting products to the newly developing organs. Many genes induced during senescence (SAGs, for senescence-associated genes) encode enzymes involved in protein degradation, underscoring the relevance of nitrogen recycling during this process [8][9][10]. Mutants exhibiting delayed leaf senescence have been described in many species and are extremely useful to identify gene products involved in cell aging, cell death and nutrient salvage [1][11][12][13][14]. While all these mutants preserve leaf greenness for extended periods, it is convenient to distinguish functional mutants, in which the delay in senescence is coupled to preservation of metabolic capacity, from those that retain green color but show normal aging behavior [15]. The latter are just defective in Chl breakdown and were categorized as non-functional stay-green mutants, also termed cosmetic [16][17].
The onset and progression of the senescence process respond to developmental cues but are also affected by environmental factors (Figure 1) [18][19]. Indeed, senescence can be induced in otherwise young leaves by darkness, abiotic stresses and microorganisms [20][21][22][23][24]. Endogenous signaling molecules and pathways, including phytohormones, reactive oxygen species (ROS) and other redox-based signals, mediate the plant responses to these inputs, which in turn lead to extensive genetic, physiological and metabolic reprogramming.
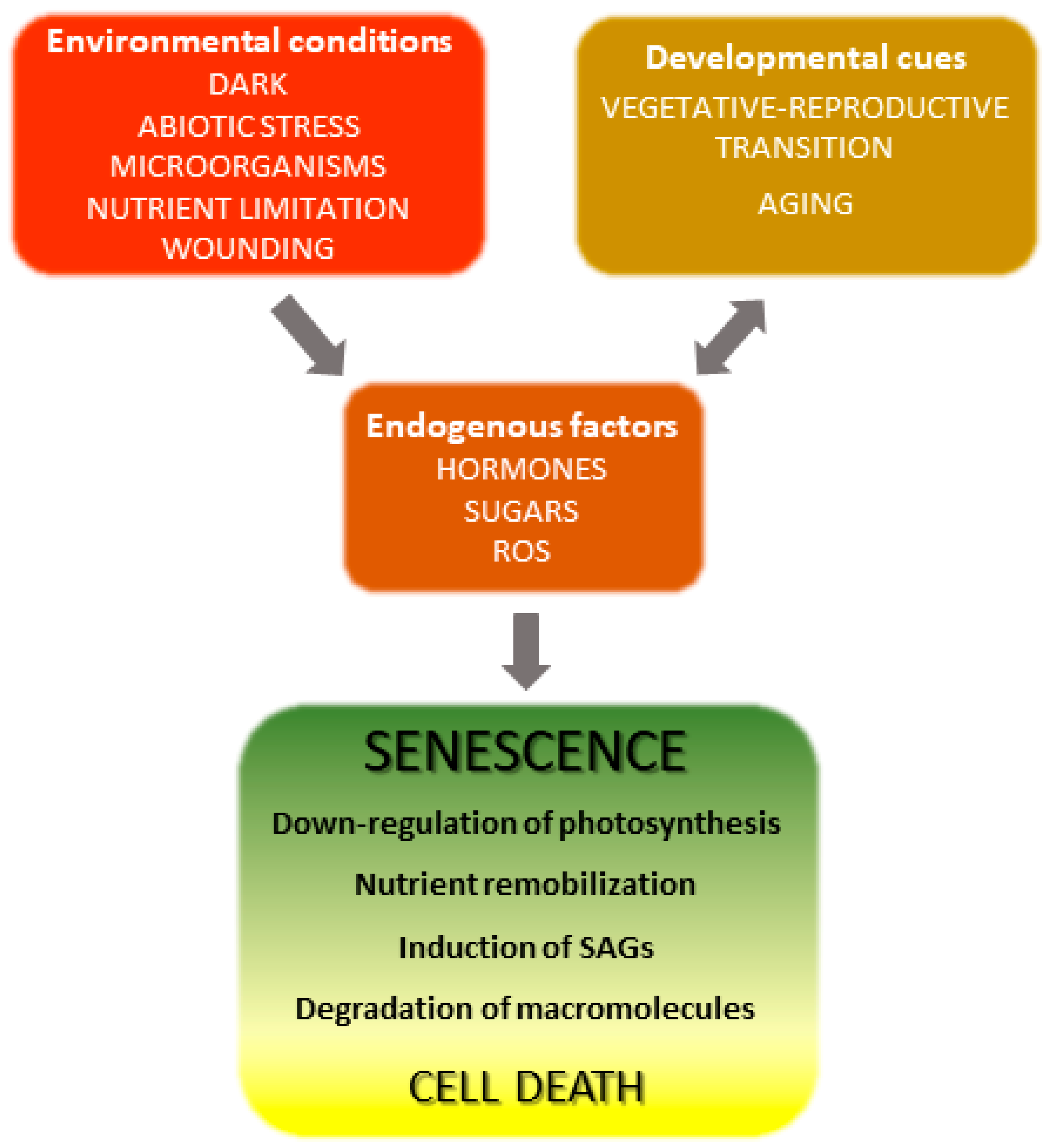 Figure 1. Overview of leaf senescence. The final stages of leaf development are basically determined by plant age and transition into the reproductive stage, but are also modulated by endogenous and exogenous cues which integrate into the developmental program. Environmental conditions affecting senescence progression include biotic and abiotic stresses and nutritional status, whereas hormones represent the most relevant endogenous factors. Many environmental stresses increase propagation of reactive oxygen species (ROS) in leaf tissue, which act as signaling molecules. SAGs, senescence-associated genes.
Figure 1. Overview of leaf senescence. The final stages of leaf development are basically determined by plant age and transition into the reproductive stage, but are also modulated by endogenous and exogenous cues which integrate into the developmental program. Environmental conditions affecting senescence progression include biotic and abiotic stresses and nutritional status, whereas hormones represent the most relevant endogenous factors. Many environmental stresses increase propagation of reactive oxygen species (ROS) in leaf tissue, which act as signaling molecules. SAGs, senescence-associated genes.Involvement of ROS such as hydrogen peroxide (H2O2), singlet oxygen (1O2) and the superoxide (O2.−) radical in both natural and induced plant senescence is supported by many observations [25][26][27][28]. ROS can be produced in various cellular compartments through the activity of oxidases or as byproducts of oxido-reductive processes (Figure 2), and the contributions of the different sources to plant senescence are still poorly understood. In animal systems, ROS associated to mitochondrial metabolism play a central role in cell aging [29]. While a similar mechanism is likely to operate in nonphotosynthetic tissues [30][31], chloroplasts are the main ROS-producing organelle in illuminated leaves, whereas peroxisomes make a substantial contribution under photorespiratory conditions in C3 plants [32][33].
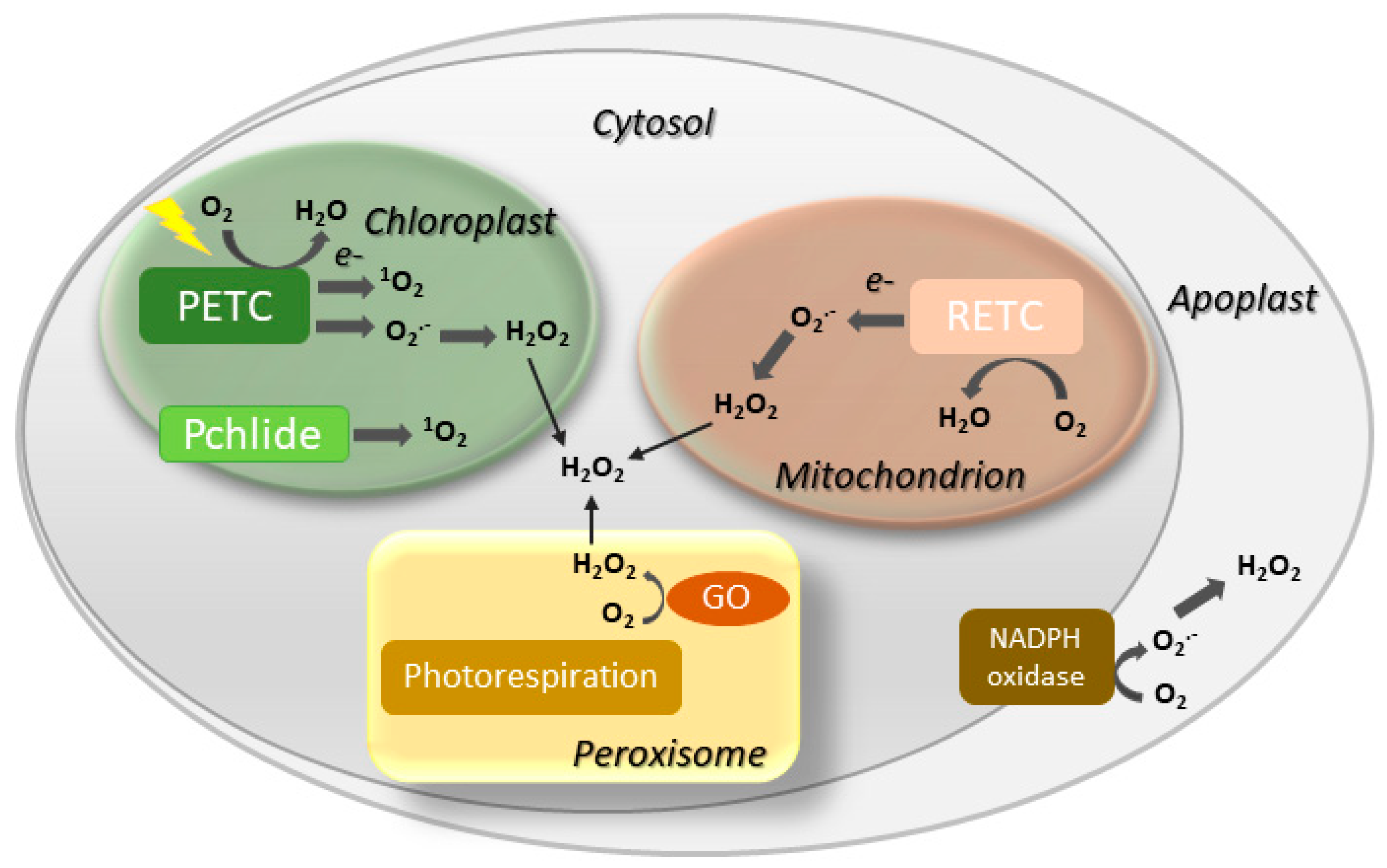 Figure 2. Major sites of ROS generation in the plant cell. GO, glycolate oxidase; Pchlide, protochlorophyllide; PETC, photosynthetic electron transport chain; RETC, respiratory electron transport chain. Disproportionation of O2.− into H2O2 may be spontaneous or mediated by a suite of superoxide dismutases.
Figure 2. Major sites of ROS generation in the plant cell. GO, glycolate oxidase; Pchlide, protochlorophyllide; PETC, photosynthetic electron transport chain; RETC, respiratory electron transport chain. Disproportionation of O2.− into H2O2 may be spontaneous or mediated by a suite of superoxide dismutases.Although the knowledge on the participation of chloroplast redox chemistry in plant senescence and programmed cell death lags behind that of animal mitochondria, an increasing number of studies indicate that plastids might be playing a more important role than thought before during leaf senescence. The aim here is to critically review the evidence that supports this connection and to identify future research trends in the area. Since leaf senescence is not only a very interesting and significant biological question [3][34], but also bears relevance for agriculture [1][35][36], understanding the molecular mechanisms that underlie this developmental process might open new avenues to increase crop yield through an extended provision of leaf photosynthates to fruits, seeds and tubers [6].
2. Leaf Senescence Is Modulated by Multiple Inputs
Several phytohormones influence leaf aging and cell death. Gibberellic acid (GA), auxins and cytokinins have been shown to delay senescence, whereas ethylene, jasmonic acid (JA), abscisic acid (ABA) and salicylic acid (SA) accelerate it (Figure 3) [18][37]. Cytokinins are able to retard senescence in plants and detached leaves, preventing Chl degradation and destruction of metabolic activity [38][39]. Conversely, a decline of the cytokinin pool is often accompanied by a decrease of photosynthetic activity and enhanced senescence. Auxins play a similar role by modulating expression of several auxin-responsive transcription factors (ARFs, Figure 3), which affect several processes associated with leaf senescence [40]. Moreover, increased expression of YUC6, a gene encoding a flavin-containing monooxygenase that catalyzes the rate-limiting step of auxin biosynthesis, was shown to delay senescence in transgenic Arabidopsis plants [41]. A different class of anti-senescence phytohormones is represented by pentacyclic diterpenes of the GA family, which act in a defined time-frame as the active GA forms are progressively degraded with leaf aging [42]. Treatment with inhibitors of GA synthesis led to increased ABA contents and promotion of senescence, suggesting that GA and ABA have antagonistic functions during this process [43].
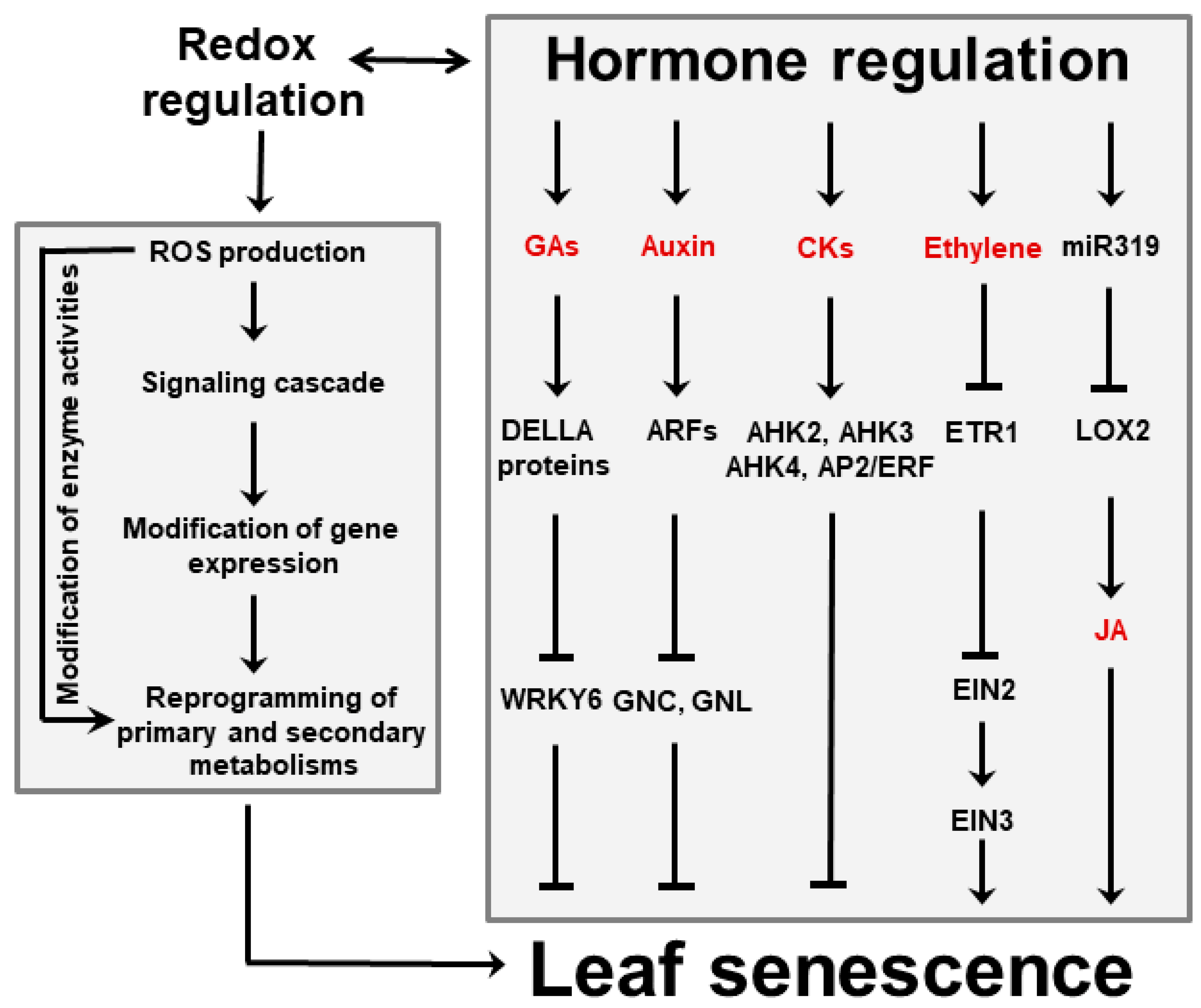 Figure 3. Interactions between hormones and ROS during leaf senescence regulation. Plant hormones can affect leaf aging either directly or via signaling cascades in which ROS are involved. Among them, GAs, auxins and CKs delay senescence, whereas ethylene and JA favor it. Regulation of JA accumulation via miR319 and lipoxygenase 2 (LOX2) is indicated. ROS might also act as independent signaling molecules that are produced upon leaf aging and stress conditions. ROS trigger modifications of gene expression that in turn result in reprogramming primary and secondary metabolisms involving sugars, energy, amino acids and antioxidants. Examples of reported interactions between hormones and ROS are given in the text. Abbreviations for hormones are also found there. AHK, Arabidopsis histidine kinase; AP2, Apetala 2; CKs, cytokinins; EIN, ethylene-insensitive protein; ERF, ethylene-responsive factor; ETR1, ethylene receptor 1; GNC, carbon-metabolism involved; GNL, GNC-like.
Figure 3. Interactions between hormones and ROS during leaf senescence regulation. Plant hormones can affect leaf aging either directly or via signaling cascades in which ROS are involved. Among them, GAs, auxins and CKs delay senescence, whereas ethylene and JA favor it. Regulation of JA accumulation via miR319 and lipoxygenase 2 (LOX2) is indicated. ROS might also act as independent signaling molecules that are produced upon leaf aging and stress conditions. ROS trigger modifications of gene expression that in turn result in reprogramming primary and secondary metabolisms involving sugars, energy, amino acids and antioxidants. Examples of reported interactions between hormones and ROS are given in the text. Abbreviations for hormones are also found there. AHK, Arabidopsis histidine kinase; AP2, Apetala 2; CKs, cytokinins; EIN, ethylene-insensitive protein; ERF, ethylene-responsive factor; ETR1, ethylene receptor 1; GNC, carbon-metabolism involved; GNL, GNC-like.Besides ABA, ethylene and JA are emerging as key players in senescence induction. They act directly or via interactions with each other, and involve various transcription factors and microRNAs (miR319, Figure 3). The relationship between ethylene and senescence has been best studied using Arabidopsis ethylene-insensitive mutants which display delayed leaf senescence [44]. Likewise, studies using Arabidopsis mutants exhibiting reduced JA levels or insensitivity to JA signaling revealed that the onset of natural and dark-induced senescence was delayed as the levels or sensitivity to this hormone declined [45][46].
ROS, phytohormones and environmental inputs do not operate through independent pathways, but instead display a significant degree of interaction and cross-talk. For instance, Merewitz et al. [47][48] have shown that higher cytokinin contents, obtained by over-expression of rate-limiting biosynthetic enzymes, resulted in induction of stress-responsive proteins such as antioxidants and chaperones, and led to improved drought tolerance. Conversely, ethylene biosynthesis during age-dependent and dark-induced leaf senescence were mediated by phytochrome-interacting transcription factors in Arabidopsis [49]. Interactions between ROS and phytohormones on the regulation of the senescence process have been demonstrated for auxins [40], GA [50][51] and SA [52]. Finally, many plant developmental programs depend on the combined action of several hormones that interact cooperatively or antagonistically. Cytokinin, in particular, has been shown to participate in regulatory networks with auxins, GA, ABA and strigolactones [53], and cross-talk between cytokinin and ethylene signaling pathways regulates leaf abscission in cotton during chemically induced senescence [54].
3. Senescence and Cell Death
The ultimate outcome of senescence is cell death and the two terms are sometimes employed indistinctly, but death is only the final stage of a complex and ordered physiological process. Indeed, there is a clear distinction between the tightly regulated senescence program that requires living cells and nutrient recycling, and the irreversible terminal phase of cell death, despite the common players and pathways they might share [7]. Similar to senescence, cell death can be induced independently of tissue age by many environmental stimuli including abiotic stress, pathogens, nutritional shortage and xenobiotics [1][55]. Biotic interactions offer a particularly rich set of examples. Biotrophic pathogens require living tissue to grow and manipulate host physiology to obtain nutrients, whereas necrotrophs kill host cells and feed on them [56][57][58]. Plants exposed to invading microorganisms often elicit a multigenic response termed the hypersensitive reaction (HR) that leads in most cases to localized cell death (LCD) at the site of infection [56]. It is assumed that the LCD associated to the HR helps to contain biotrophs by opposing a barrier of dead cells which deter their advance into the adjacent living tissue [56]. Infection by necrotrophic pathogens, instead, is facilitated by cell death and some necrotrophs may even promote host LCD in their own benefit [57].
As in the case of senescence, increased ROS production is a quasi-universal feature of cell death induced by environmental stresses and the HR [59][60][61][62][63]. In leaves, a significant fraction of all ROS are generated as byproducts of photosynthetic electron transport (Figure 2). Partial reduction of oxygen at the level of photosystem (PS) I and PSII renders O2.− and, after spontaneous or enzyme-mediated disproportionation, H2O2 [64][65]. Singlet oxygen is usually produced in PSII by energy transfer from excited triplet-state Chl moieties to basal triplet-state oxygen resulting in spin inversion and several-fold increase in reactivity [66]. Under normal photosynthetic conditions, ROS build-up is limited by the action of antioxidant enzymes and reductants, but whenever distribution of reducing equivalents is perturbed by biotic or abiotic stresses, the rate of leakage increases dramatically, overcoming the control devices and leading to ROS propagation [67][68].
Another major source of chloroplast ROS is Chl metabolism (Figure 2). Many biosynthetic intermediates are loosely bound to the thylakoids without associating with reaction centers or antenna [69]. They are unable to participate in photosynthesis but can still be excited to their triplet state in the light, readily reacting with oxygen and propagating 1O2. Chl breakdown products released from the photosystems can also engage in this energy transfer reaction [5], and have been proposed to promote cell death during pathogen-induced HR [70].
The first report linking chloroplast redox processes to cell death came from the research of Samuilov et al. [71], who observed that cyanide treatment caused light-dependent destruction of chloroplast-containing leaf guard cells, whereas heterotrophic epidermal tissue was not affected. Photosynthesis-associated events such as over-reduction of plastoquinone and ROS build-up were proposed to mediate this effect [71]. Heat-induced cell death has also been linked to chloroplast redox chemistry [72][73]. A complete death program is triggered by 1O2 via the plastidic proteins Executioner 1 and 2, as revealed by experiments on the Arabidopsis flu mutants, which accumulate the photosensitive biosynthetic intermediate protochlorophyllide in the dark and propagates 1O2 upon illumination (Figure 2; [74]). Moreover, cell death associated to the HR is significantly reduced in the absence of light [75][76], and can be abolished by targeting antioxidant proteins to chloroplasts [77]. In addition to these examples, many other reports identified chloroplast ROS as key players in the signaling events that lead to programmed cell death [69][78][79][80]. In contrast, little is known on the nature of the death-promoting signals that exit the chloroplast, and the relationship of the cell death executioners with the organelle.
References
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622.
- Zhang, H.; Zhou, C. Signal transduction in leaf senescence. Plant Mol. Biol. 2013, 82, 539–545.
- Schippers, J.H.; Schmidt, R.; Wagstaff, C.; Jing, H.-C. Living to die and dying to live: The survival strategy behind leaf senescence. Plant Physiol. 2015, 169, 914–930.
- Tamary, E.; Nevo, R.; Naveh, L.; Levin-Zaidman, S.; Kiss, V.; Savidor, A.; Levin, Y.; Eyal, Y.; Reich, Z.; Adam, Z. Chlorophyll catabolism precedes changes in chloroplast structure and proteome during leaf senescence. Plant Direct 2019, 3, 1–18.
- Hörtensteiner, S.; Kräutler, B. Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta 2011, 1807, 977–988.
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf senescence: Systems and dynamics aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376.
- Thomas, H. Senescence, ageing and death of the whole plant. New Phytol. 2013, 197, 696–711.
- Buet, A.; Costa, M.L.; Martínez, D.E.; Guiamet, J.J. Chloroplast protein degradation in senescing leaves: Proteases and lytic compartments. Front. Plant Sci. 2019, 10, 747.
- Uzelac, B.; Janošević, D.; Simonović, A.; Motyka, V.; Dobrev, P.I.; Budimir, S. Characterization of natural leaf senescence in tobacco (Nicotiana tabacum) plants grown in vitro. Protoplasma 2016, 253, 259–275.
- Zentgraf, U. Tug-of-war during senescence. Nat. Plants 2019, 5, 129–130.
- Thomas, H.; Ougham, H.; Canter, P.; Donnison, I. What stay-green mutants tell us about nitrogen remobilization in leaf senescence. J. Exp. Bot. 2002, 53, 801–808.
- Ramkumar, M.; Senthil Kumar, S.; Gaikwad, K.; Pandey, R.; Chinnusamy, V.; Singh, N.K.; Singh, A.K.; Mohapatra, T.; Sevanthi, A.M. A novel stay-green mutant of rice with delayed leaf senescence and better harvest index confers drought tolerance. Plants 2019, 8, 375.
- Sekhon, R.S.; Saski, C.; Kumar, R.; Flinn, B.S.; Luo, F.; Beissinger, T.M.; Ackerman, A.J.; Breitzman, M.W.; Bridges, W.C.; de Leon, N. Integrated genome-scale analysis identifies novel genes and networks underlying senescence in maize. Plant Cell 2019, 31, 1968–1989.
- Wang, J.; Huang, R. Modulation of ethylene and ascorbic acid on reactive oxygen species scavenging in plant salt response. Front. Plant Sci. 2019, 10, 319.
- Thomas, H.; Howarth, C.J. Five ways to stay green. J. Exp. Bot. 2000, 51, 329–337.
- Thomas, H.; Ougham, H. The stay-green trait. J. Exp. Bot. 2014, 65, 3889–3900.
- Myers, J.R.; Aljadi, M.; Brewer, L. The importance of cosmetic stay-green in specialty crops. In Plant Breeding Reviews; Goldman, I., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; Volume 42, pp. 219–256.
- Jibran, R.; Hunter, D.A.; Dijkwel, P.P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013, 82, 547–561.
- Ay, N.; Janack, B.; Humbeck, K. Epigenetic control of plant senescence and linked processes. J. Exp. Bot. 2014, 65, 3875–3887.
- Häffner, E.; Konietzki, S.; Diederichsen, E. Keeping control: The role of senescence and development in plant pathogenesis and defense. Plants 2015, 4, 449–488.
- Bhattacharjee, S. ROS in aging and senescence. In Reactive Oxygen Species in Plant Biology; Springer: New Delhi, India, 2019; pp. 65–79.
- Shi, X.; Xu, S.; Mu, D.; Sadeghnezhad, E.; Li, Q.; Ma, Z.; Zhao, L.; Zhang, Q.; Wang, L. Exogenous melatonin delays dark-induced grape leaf senescence by regulation of antioxidant system and senescence associated genes (SAGs). Plants 2019, 8, 366.
- Sade, N.; del Mar Rubio-Wilhelmi, M.; Umnajkitikorn, K.; Blumwald, E. Stress-induced senescence and plant tolerance to abiotic stress. J. Exp. Bot. 2017, 69, 845–853.
- Liu, W.; Zhang, W.; Zheng, N.; Zhai, W.; Qi, F. Study of cotton leaf senescence induced by Alternaria alternata infection. In Plant Senescence, Methods in Molecular Biology; Guo, Y., Ed.; Humana Press: New York, NY, USA, 2018; Volume 1744, pp. 161–171.
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.-I.; Asensi-Fabado, M.A.; Munné-Bosch, S.; Antonio, C.; Tohge, T. JUNGBRUNNEN1, a reactive oxygen species–responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506.
- Xie, Y.; Huhn, K.; Brandt, R.; Potschin, M.; Bieker, S.; Straub, D.; Doll, J.; Drechsler, T.; Zentgraf, U.; Wenkel, S. REVOLUTA and WRKY53 connect early and late leaf development in Arabidopsis. Development 2014, 141, 4772–4783.
- Biswas, M.S.; Mano, J.I. Lipid peroxide-derived short-chain carbonyls mediate hydrogen peroxide-induced and salt-induced programmed cell death in plants. Plant Physiol. 2015, 168, 885–898.
- Garapati, P.; Xue, G.-P.; Munné-Bosch, S.; Balazadeh, S. Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 2015, 168, 1122–1139.
- Circu, M.L.; Aw, T.Y. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Biol. Med. 2010, 48, 749–762.
- Rogers, H.; Munné-Bosch, S. Production and scavenging of reactive oxygen species and redox signaling during leaf and flower senescence: Similar but different. Plant Physiol. 2016, 171, 1560–1568.
- Muñoz, P.; Munné-Bosch, S. Photo-oxidative stress during leaf, flower and fruit development. Plant Physiol. 2018, 176, 1004–1014.
- Wang, S.; Blumwald, E. Stress-induced chloroplast degradation in Arabidopsis is regulated via a process independent of autophagy and senescence-associated vacuoles. Plant Cell 2014, 26, 4875–4888.
- Noctor, G.; Foyer, C.H. Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 2016, 171, 1581–1592.
- Penfold, C.A.; Buchanan-Wollaston, V. Modelling transcriptional networks in leaf senescence. J. Exp. Bot. 2014, 65, 3859–3873.
- Antonietta, M.; Acciaresi, H.; Guiamet, J. Responses to N deficiency in stay green and non-stay green argentinean hybrids of maize. J. Agron. Crop Sci. 2016, 202, 231–242.
- Moschen, S.; Higgins, J.; Di Rienzo, J.A.; Heinz, R.A.; Paniego, N.; Fernández, P. Network and biosignature analysis for the integration of transcriptomic and metabolomic data to characterize leaf senescence process in sunflower. BMC Bioinform. 2016, 17, 174.
- Jan, S.; Abbas, N.; Ashraf, M.; Ahmad, P. Roles of potential plant hormones and transcription factors in controlling leaf senescence and drought tolerance. Protoplasma 2019, 256, 313–329.
- Talla, S.K.; Panigrahy, M.; Kappara, S.; Nirosha, P.; Neelamraju, S.; Ramanan, R. Cytokinin delays dark-induced senescence in rice by maintaining the chlorophyll cycle and photosynthetic complexes. J. Exp. Bot. 2016, 67, 1839–1851.
- Lu, G.; Casaretto, J.A.; Ying, S.; Mahmood, K.; Liu, F.; Bi, Y.-M.; Rothstein, S.J. Overexpression of OsGATA12 regulates chlorophyll content, delays plant senescence and improves rice yield under high density planting. Plant Mol. Biol. 2017, 94, 215–227.
- Mueller-Roeber, B.; Balazadeh, S. Auxin and its role in plant senescence. J. Plant Growth Regul. 2014, 33, 21–33.
- Kim, J.I.; Murphy, A.S.; Baek, D.; Lee, S.-W.; Yun, D.-J.; Bressan, R.A.; Narasimhan, M.L. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 3981–3992.
- van der Graaff, E.; Schwacke, R.; Schneider, A.; Desimone, M.; Flügge, U.-I.; Kunze, R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006, 141, 776–792.
- Yu, K.; Wei, J.; Ma, Q.; Yu, D.; Li, J. Senescence of aerial parts is impeded by exogenous gibberellic acid in herbaceous perennial Paris polyphylla. J. Plant Physiol. 2009, 166, 819–830.
- Kim, J.; Chang, C.; Tucker, M.L. To grow old: Regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 2015, 6, 20.
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230.
- He, Y.; Gan, S. A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 2002, 14, 805–815.
- Merewitz, E.B.; Gianfagna, T.; Huang, B. Photosynthesis, water use, and root viability under water stress as affected by expression of SAG12-ipt controlling cytokinin synthesis in Agrostis stolonifera. J. Exp. Bot. 2011, 62, 383–395.
- Merewitz, E.; Xu, Y.; Huang, B. Differentially expressed genes associated with improved drought tolerance in creeping bentgrass overexpressing a gene for cytokinin biosynthesis. PLoS ONE 2016, 11, e0166676.
- Song, Y.; Yang, C.; Gao, S.; Zhang, W.; Li, L.; Kuai, B. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787.
- Rosenwasser, S.; Belausov, E.; Riov, J.; Holdengreber, V.; Friedman, H. Gibberellic acid (GA3) inhibits ROS increase in chloroplasts during dark-induced senescence of pelargonium cuttings. J. Plant Growth Regul. 2010, 29, 375–384.
- Jajic, I.; Sarna, T.; Strzalka, K. Senescence, stress, and reactive oxygen species. Plants 2015, 4, 393–411.
- Guo, P.; Li, Z.; Huang, P.; Li, B.; Fang, S.; Chu, J.; Guo, H. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 2017, 29, 2854–2870.
- El-Showk, S.; Ruonala, R.; Helariutta, Y. Crossing paths: Cytokinin signalling and crosstalk. Development 2013, 140, 1373–1383.
- Xu, J.; Chen, L.; Sun, H.; Wusiman, N.; Sun, W.; Li, B.; Gao, Y.; Kong, J.; Zhang, D.; Zhang, X. Crosstalk between cytokinin and ethylene signaling pathways regulates leaf abscission in cotton in response to chemical defoliants. J. Exp. Bot. 2019, 70, 1525–1538.
- Gepstein, S.; Glick, B.R. Strategies to ameliorate abiotic stress-induced plant senescence. Plant Mol. Biol. 2013, 82, 623–633.
- Senthil-Kumar, M.; Mysore, K.S. Nonhost resistance against bacterial pathogens: Retrospectives and prospects. Annu. Rev. Phytopathol. 2013, 51, 407–427.
- Rossi, F.R.; Krapp, A.R.; Bisaro, F.; Maiale, S.J.; Pieckenstain, F.L.; Carrillo, N. Reactive oxygen species generated in chloroplasts contribute to tobacco leaf infection by the necrotrophic fungus Botrytis cinerea. Plant J. 2017, 92, 761–773.
- Pierella Karlusich, J.J.; Zurbriggen, M.D.; Shahinnia, F.; Sonnewald, S.; Sonnewald, U.; Hosseini, S.A.; Hajirezaei, M.-R.; Carrillo, N. Chloroplast redox status modulates genome-wide plant responses during the non-host interaction of tobacco with the hemibiotrophic bacterium Xanthomonas campestris pv. vesicatoria. Front. Plant Sci. 2017, 8, 1158.
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69.
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1092.
- Jwa, N.-S.; Hwang, B.K. Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front. Plant Sci. 2017, 8, 1687.
- Suzuki, N.; Katano, K. Coordination between ROS regulatory systems and other pathways under heat stress and pathogen attack. Front. Plant Sci. 2018, 9, 490.
- Su, J.; Yang, L.; Zhu, Q.; Wu, H.; He, Y.; Liu, Y.; Xu, J.; Jiang, D.; Zhang, S. Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector-triggered immunity. PLoS Biol. 2018, 16, e2004122.
- Kozuleva, M.A.; Ivanov, B.N. The mechanisms of oxygen reduction in the terminal reducing segment of the chloroplast photosynthetic electron transport chain. Plant Cell Physiol. 2016, 57, 1397–1404.
- Pospíšil, P. Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front. Plant Sci. 2016, 7, 1950.
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53.
- Mignolet-Spruyt, L.; Xu, E.; Idänheimo, N.; Hoeberichts, F.A.; Mühlenbock, P.; Brosché, M.; Van Breusegem, F.; Kangasjärvi, J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016, 67, 3831–3844.
- Sewelam, N.; Kazan, K.; Schenk, P.M. Global plant stress signaling: Reactive oxygen species at the cross-road. Front. Plant Sci. 2016, 7, 187.
- Ambastha, V.; Tripathy, B.C.; Tiwari, B.S. Programmed cell death in plants: A chloroplastic connection. Plant Signal. Behav. 2015, 10, e989752.
- Mur, L.A.; Aubry, S.; Mondhe, M.; Kingston-Smith, A.; Gallagher, J.; Timms-Taravella, E.; James, C.; Papp, I.; Hörtensteiner, S.; Thomas, H. Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. New Phytol. 2010, 188, 161–174.
- Samuilov, V.D.; Lagunova, E.M.; Kiselevsky, D.B.; Dzyubinskaya, E.V.; Makarova, Y.V.; Gusev, M.V. Participation of chloroplasts in plant apoptosis. Biosci. Rep. 2003, 23, 103–117.
- Doyle, S.M.; Diamond, M.; McCabe, P.F. Chloroplast and reactive oxygen species involvement in apoptotic-like programmed cell death in Arabidopsis suspension cultures. J. Exp. Bot. 2009, 61, 473–482.
- Wang, Q.-L.; Chen, J.-H.; He, N.-Y.; Guo, F.-Q. Metabolic reprogramming in chloroplasts under heat stress in plants. Int. J. Mol. Sci. 2018, 19, 849.
- Lee, K.P.; Kim, C.; Landgraf, F.; Apel, K. EXECUTER1-and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 10270–10275.
- Karpinski, S.; Gabrys, H.; Mateo, A.; Karpinska, B.; Mullineaux, P.M. Light perception in plant disease defence signalling. Curr. Opin. Plant Biol. 2003, 6, 390–396.
- Liu, Y.; Ren, D.; Pike, S.; Pallardy, S.; Gassmann, W.; Zhang, S. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 2007, 51, 941–954.
- Zurbriggen, M.D.; Carrillo, N.; Tognetti, V.B.; Melzer, M.; Peisker, M.; Hause, B.; Hajirezaei, M.-R. Chloroplast-generated reactive oxygen species play a major role in localized cell death during the non-host interaction between tobacco and Xanthomonas campestris pv. vesicatoria. Plant J. 2009, 60, 962–973.
- Van Aken, O.; Van Breusegem, F. Licensed to kill: Mitochondria, chloroplasts, and cell death. Trends Plant Sci. 2015, 20, 754–766.
- Gutiérrez, J.; González-Pérez, S.; García-García, F.; Daly, C.T.; Lorenzo, Ó.; Revuelta, J.L.; McCabe, P.F.; Arellano, J.B. Programmed cell death activated by Rose Bengal in Arabidopsis thaliana cell suspension cultures requires functional chloroplasts. J. Exp. Bot. 2014, 65, 3081–3095.
- Kim, C.; Meskauskiene, R.; Zhang, S.; Lee, K.P.; Ashok, M.L.; Blajecka, K.; Herrfurth, C.; Feussner, I.; Apel, K. Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 2012, 24, 3026–3039.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.7K
Entry Collection:
Environmental Sciences
Revisions:
3 times
(View History)
Update Date:
02 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




