Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Néstor Carrillo and Version 3 by Camila Xu.
Leaf senescence is an ordered physiological process in which cellular structures and biomolecules are progressively broken down and the resulting products mobilized to other plant organs such as fruits, seeds, tubers and/or more apical leaves.
- senescence
- reactive oxygen species
- chloroplast
- phytohormones
1. Introduction
Leaf senescence is an ordered physiological process in which cellular structures and biomolecules are progressively broken down and the resulting products mobilized to other plant organs such as fruits, seeds, tubers and/or more apical leaves [1][2][3][1,2,3]. The most visible manifestation of leaf senescence is yellowing caused by destruction of the chloroplast pigment-protein complexes and conversion of the constituent chlorophylls (Chl) into catabolic non-green derivatives after opening of the chlorin ring system [4][5][4,5]. Natural leaf senescence as it occurs in the field is normally age-dependent and accelerates upon transition of the vegetative into the reproductive growth phase [6][7][6,7].
Senescence is controlled by a genetic program involving major changes in expression patterns that result in degradation of cells targeted for demise and reallocation of the resulting products to the newly developing organs. Many genes induced during senescence (SAGs, for senescence-associated genes) encode enzymes involved in protein degradation, underscoring the relevance of nitrogen recycling during this process [8][9][10][8,9,10]. Mutants exhibiting delayed leaf senescence have been described in many species and are extremely useful to identify gene products involved in cell aging, cell death and nutrient salvage [1][11][12][13][14][1,11,12,13,14]. While all these mutants preserve leaf greenness for extended periods, it is convenient to distinguish functional mutants, in which the delay in senescence is coupled to preservation of metabolic capacity, from those that retain green color but show normal aging behavior [15]. The latter are just defective in Chl breakdown and were categorized as non-functional stay-green mutants, also termed cosmetic [16][17][16,17].
The onset and progression of the senescence process respond to developmental cues but are also affected by environmental factors (Figure 1) [18][19][18,19]. Indeed, senescence can be induced in otherwise young leaves by darkness, abiotic stresses and microorganisms [20][21][22][23][24][20,21,22,23,24]. Endogenous signaling molecules and pathways, including phytohormones, reactive oxygen species (ROS) and other redox-based signals, mediate the plant responses to these inputs, which in turn lead to extensive genetic, physiological and metabolic reprogramming.
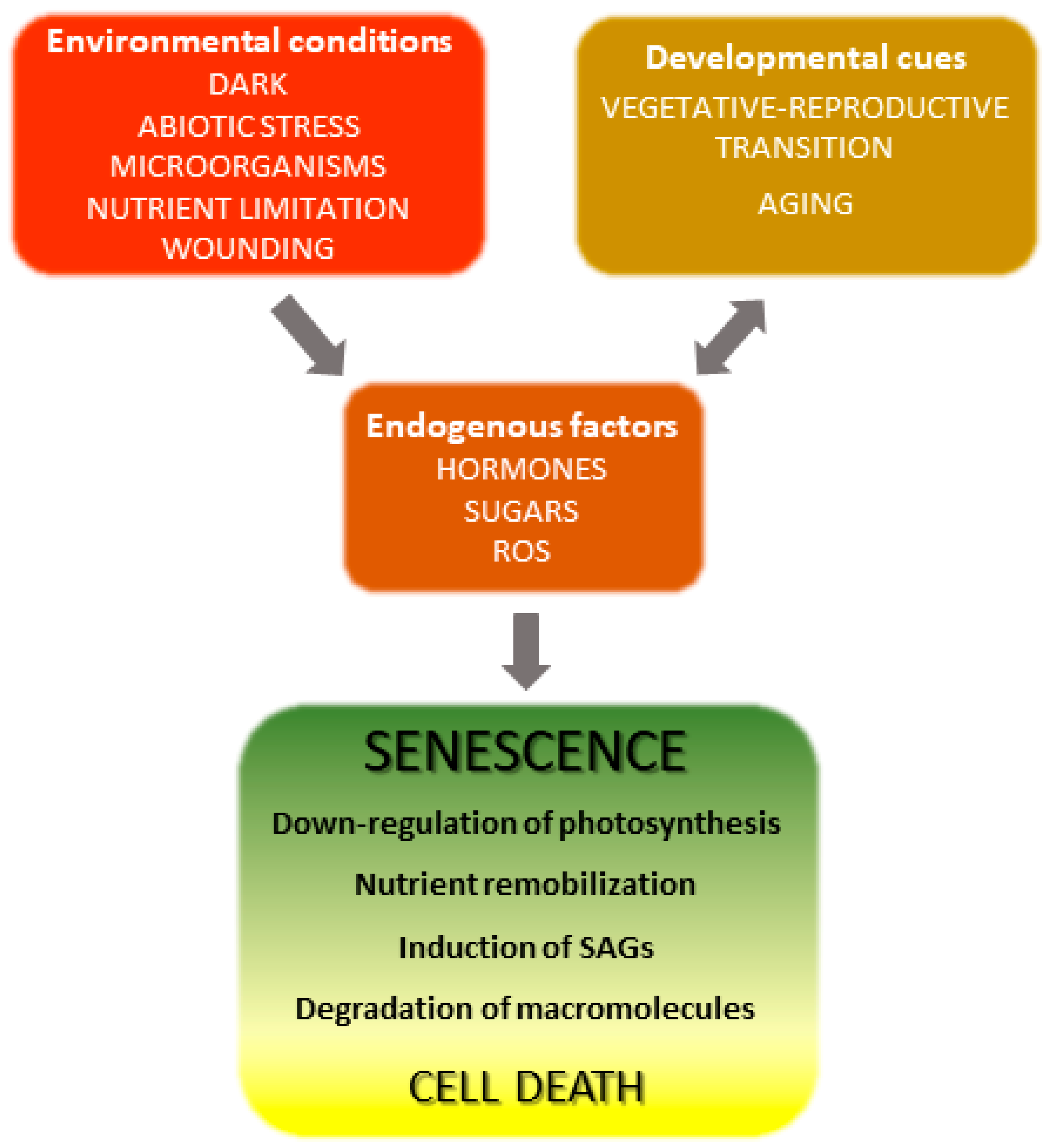
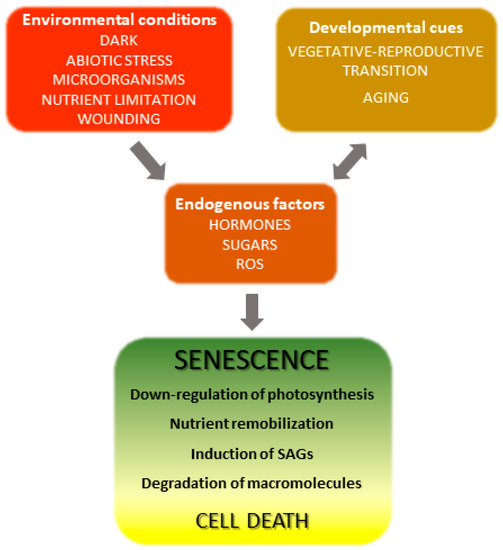
Figure 1. Overview of leaf senescence. The final stages of leaf development are basically determined by plant age and transition into the reproductive stage, but are also modulated by endogenous and exogenous cues which integrate into the developmental program. Environmental conditions affecting senescence progression include biotic and abiotic stresses and nutritional status, whereas hormones represent the most relevant endogenous factors. Many environmental stresses increase propagation of reactive oxygen species (ROS) in leaf tissue, which act as signaling molecules. SAGs, senescence-associated genes.
Involvement of ROS such as hydrogen peroxide (H2O2), singlet oxygen (1O2) and the superoxide (O2.−) radical in both natural and induced plant senescence is supported by many observations [25][26][27][28][25,26,27,28]. ROS can be produced in various cellular compartments through the activity of oxidases or as byproducts of oxido-reductive processes (Figure 2), and the contributions of the different sources to plant senescence are still poorly understood. In animal systems, ROS associated to mitochondrial metabolism play a central role in cell aging [29]. While a similar mechanism is likely to operate in nonphotosynthetic tissues [30][31][30,31], chloroplasts are the main ROS-producing organelle in illuminated leaves, whereas peroxisomes make a substantial contribution under photorespiratory conditions in C3 plants [32][33][32,33].
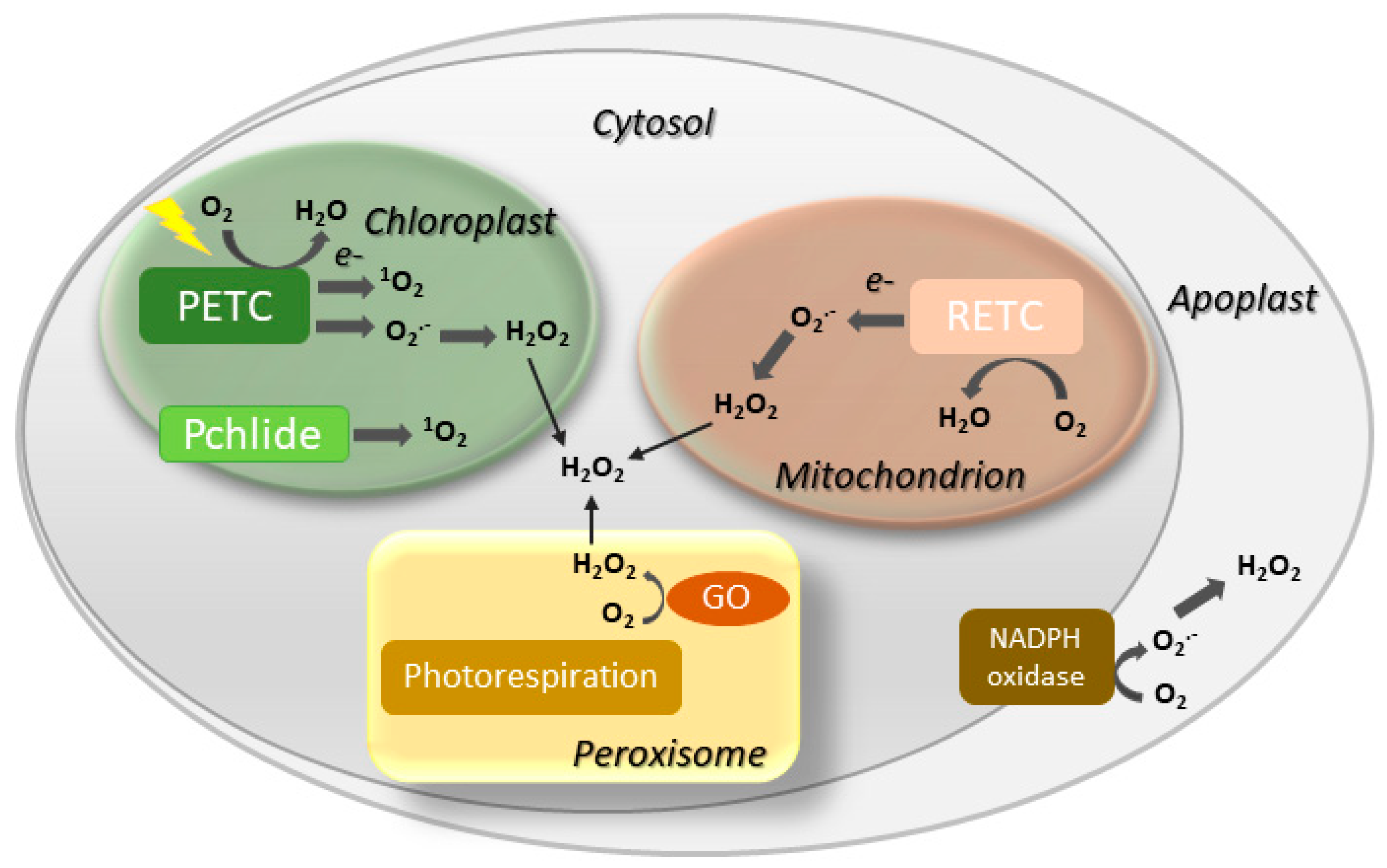
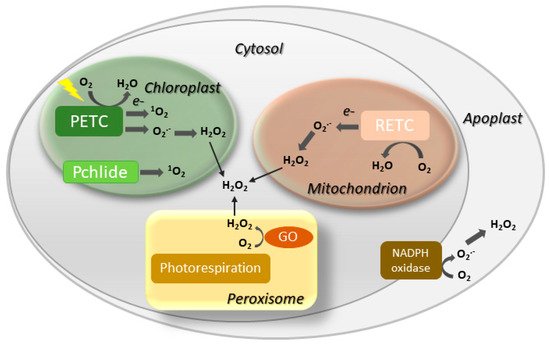
Figure 2. Major sites of ROS generation in the plant cell. GO, glycolate oxidase; Pchlide, protochlorophyllide; PETC, photosynthetic electron transport chain; RETC, respiratory electron transport chain. Disproportionation of O2.− into H2O2 may be spontaneous or mediated by a suite of superoxide dismutases.
Although theour knowledge on the participation of chloroplast redox chemistry in plant senescence and programmed cell death lags behind that of animal mitochondria, an increasing number of studies indicate that plastids might be playing a more important role than thought before during leaf senescence. The aim her of this article is to critically review the evidence that supports this connection and to identify future research trends in the area. Since leaf senescence is not only a very interesting and significant biological question [3][34][3,34], but also bears relevance for agriculture [1][35][36][1,35,36], understanding the molecular mechanisms that underlie this developmental process might open new avenues to increase crop yield through an extended provision of leaf photosynthates to fruits, seeds and tubers [6].
2. Leaf Senescence Is Modulated by Multiple Inputs
Several phytohormones influence leaf aging and cell death. Gibberellic acid (GA), auxins and cytokinins have been shown to delay senescence, whereas ethylene, jasmonic acid (JA), abscisic acid (ABA) and salicylic acid (SA) accelerate it (Figure 3) [18][37][18,37]. Cytokinins are able to retard senescence in plants and detached leaves, preventing Chl degradation and destruction of metabolic activity [38][39][38,39]. Conversely, a decline of the cytokinin pool is often accompanied by a decrease of photosynthetic activity and enhanced senescence. Auxins play a similar role by modulating expression of several auxin-responsive transcription factors (ARFs, Figure 3), which affect several processes associated with leaf senescence [40]. Moreover, increased expression of YUC6, a gene encoding a flavin-containing monooxygenase that catalyzes the rate-limiting step of auxin biosynthesis, was shown to delay senescence in transgenic Arabidopsis plants [41]. A different class of anti-senescence phytohormones is represented by pentacyclic diterpenes of the GA family, which act in a defined time-frame as the active GA forms are progressively degraded with leaf aging [42]. Treatment with inhibitors of GA synthesis led to increased ABA contents and promotion of senescence, suggesting that GA and ABA have antagonistic functions during this process [43].
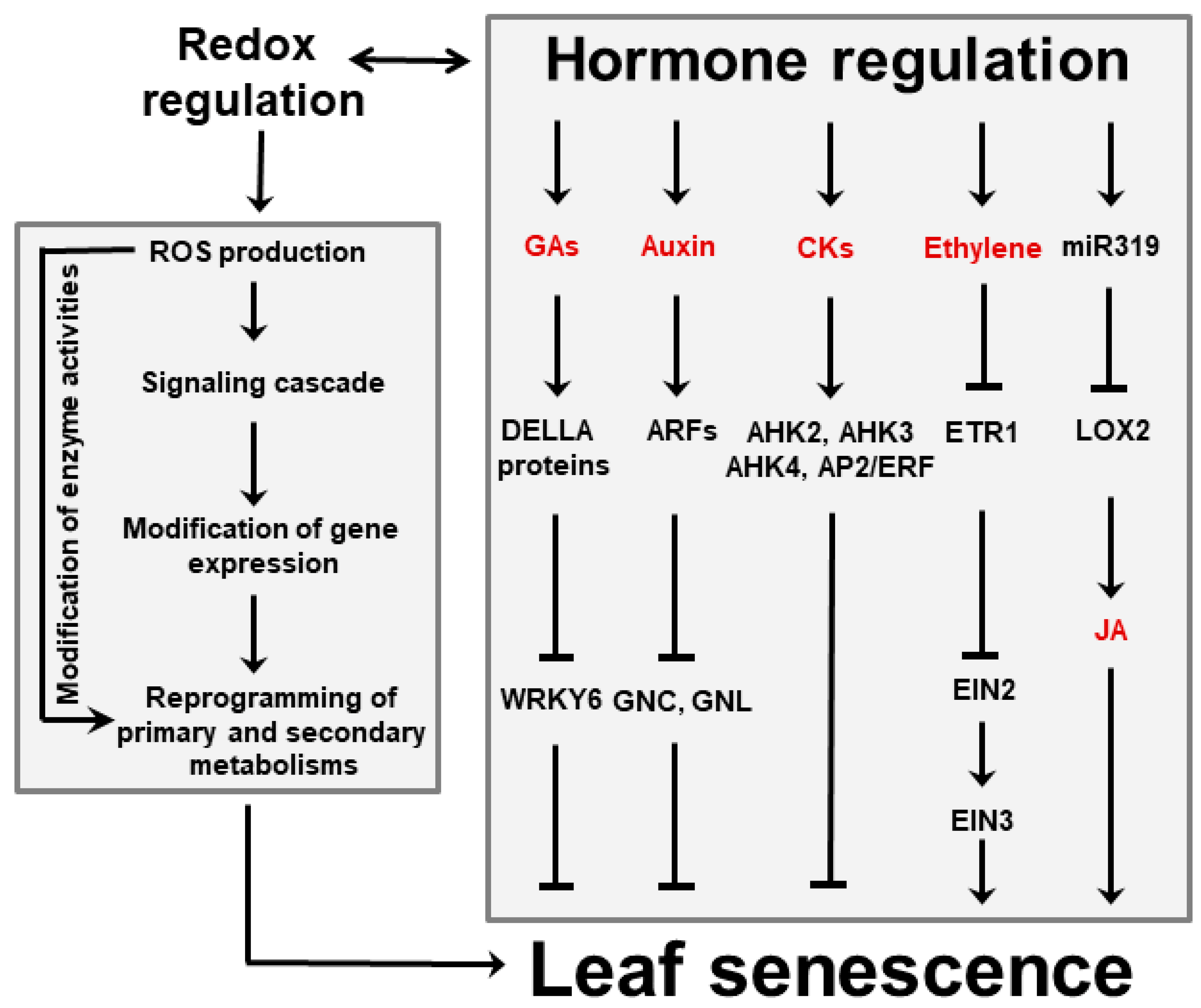
Figure 3. Interactions between hormones and ROS during leaf senescence regulation. Plant hormones can affect leaf aging either directly or via signaling cascades in which ROS are involved. Among them, GAs, auxins and CKs delay senescence, whereas ethylene and JA favor it. Regulation of JA accumulation via miR319 and lipoxygenase 2 (LOX2) is indicated. ROS might also act as independent signaling molecules that are produced upon leaf aging and stress conditions. ROS trigger modifications of gene expression that in turn result in reprogramming primary and secondary metabolisms involving sugars, energy, amino acids and antioxidants. Examples of reported interactions between hormones and ROS are given in the text. Abbreviations for hormones are also found there. AHK, Arabidopsis histidine kinase; AP2, Apetala 2; CKs, cytokinins; EIN, ethylene-insensitive protein; ERF, ethylene-responsive factor; ETR1, ethylene receptor 1; GNC, carbon-metabolism involved; GNL, GNC-like.
Besides ABA, ethylene and JA are emerging as key players in senescence induction. They act directly or via interactions with each other, and involve various transcription factors and microRNAs (miR319, Figure 3). The relationship between ethylene and senescence has been best studied using Arabidopsis ethylene-insensitive mutants which display delayed leaf senescence [44]. Likewise, studies using Arabidopsis mutants exhibiting reduced JA levels or insensitivity to JA signaling revealed that the onset of natural and dark-induced senescence was delayed as the levels or sensitivity to this hormone declined [45][46][45,46].
ROS, phytohormones and environmental inputs do not operate through independent pathways, but instead display a significant degree of interaction and cross-talk. For instance, Merewitz et al. [47][48][47,48] have shown that higher cytokinin contents, obtained by over-expression of rate-limiting biosynthetic enzymes, resulted in induction of stress-responsive proteins such as antioxidants and chaperones, and led to improved drought tolerance. Conversely, ethylene biosynthesis during age-dependent and dark-induced leaf senescence were mediated by phytochrome-interacting transcription factors in Arabidopsis [49]. Interactions between ROS and phytohormones on the regulation of the senescence process have been demonstrated for auxins [40], GA [50][51][50,51] and SA [52]. Finally, many plant developmental programs depend on the combined action of several hormones that interact cooperatively or antagonistically. Cytokinin, in particular, has been shown to participate in regulatory networks with auxins, GA, ABA and strigolactones [53], and cross-talk between cytokinin and ethylene signaling pathways regulates leaf abscission in cotton during chemically induced senescence [54].
3. Senescence and Cell Death
The ultimate outcome of senescence is cell death and the two terms are sometimes employed indistinctly, but death is only the final stage of a complex and ordered physiological process. Indeed, there is a clear distinction between the tightly regulated senescence program that requires living cells and nutrient recycling, and the irreversible terminal phase of cell death, despite the common players and pathways they might share [7]. Similar to senescence, cell death can be induced independently of tissue age by many environmental stimuli including abiotic stress, pathogens, nutritional shortage and xenobiotics [1][55][1,55]. Biotic interactions offer a particularly rich set of examples. Biotrophic pathogens require living tissue to grow and manipulate host physiology to obtain nutrients, whereas necrotrophs kill host cells and feed on them [56][57][58][56,57,58]. Plants exposed to invading microorganisms often elicit a multigenic response termed the hypersensitive reaction (HR) that leads in most cases to localized cell death (LCD) at the site of infection [56]. It is assumed that the LCD associated to the HR helps to contain biotrophs by opposing a barrier of dead cells which deter their advance into the adjacent living tissue [56]. Infection by necrotrophic pathogens, instead, is facilitated by cell death and some necrotrophs may even promote host LCD in their own benefit [57].
As in the case of senescence, increased ROS production is a quasi-universal feature of cell death induced by environmental stresses and the HR [59][60][61][62][63][59,60,61,62,63]. In leaves, a significant fraction of all ROS are generated as byproducts of photosynthetic electron transport (Figure 2). Partial reduction of oxygen at the level of photosystem (PS) I and PSII renders O2.− and, after spontaneous or enzyme-mediated disproportionation, H2O2 [64][65][64,65]. Singlet oxygen is usually produced in PSII by energy transfer from excited triplet-state Chl moieties to basal triplet-state oxygen resulting in spin inversion and several-fold increase in reactivity [66]. Under normal photosynthetic conditions, ROS build-up is limited by the action of antioxidant enzymes and reductants, but whenever distribution of reducing equivalents is perturbed by biotic or abiotic stresses, the rate of leakage increases dramatically, overcoming the control devices and leading to ROS propagation [67][68][67,68].
Another major source of chloroplast ROS is Chl metabolism (Figure 2). Many biosynthetic intermediates are loosely bound to the thylakoids without associating with reaction centers or antenna [69]. They are unable to participate in photosynthesis but can still be excited to their triplet state in the light, readily reacting with oxygen and propagating 1O2. Chl breakdown products released from the photosystems can also engage in this energy transfer reaction [5], and have been proposed to promote cell death during pathogen-induced HR [70].
The first report linking chloroplast redox processes to cell death came from the research of Samuilov et al. [71], who observed that cyanide treatment caused light-dependent destruction of chloroplast-containing leaf guard cells, whereas heterotrophic epidermal tissue was not affected. Photosynthesis-associated events such as over-reduction of plastoquinone and ROS build-up were proposed to mediate this effect [71]. Heat-induced cell death has also been linked to chloroplast redox chemistry [72][73][72,73]. A complete death program is triggered by 1O2 via the plastidic proteins Executioner 1 and 2, as revealed by experiments on the Arabidopsis flu mutants, which accumulate the photosensitive biosynthetic intermediate protochlorophyllide in the dark and propagates 1O2 upon illumination (Figure 2; [74]). Moreover, cell death associated to the HR is significantly reduced in the absence of light [75][76][75,76], and can be abolished by targeting antioxidant proteins to chloroplasts [77]. In addition to these examples, many other reports identified chloroplast ROS as key players in the signaling events that lead to programmed cell death [69][78][79][80][69,78,79,80]. In contrast, little is known on the nature of the death-promoting signals that exit the chloroplast, and the relationship of the cell death executioners with the organelle.
[1][2][3][4][5][6][7][8][9][10][11][12][13][14][15][5][16][17][18][19][5][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][5][67][68][69][70][5][71][72][73][74][75][76][77][78][79][80][81][82][83][84][85][86][87][88][89][90][91][92][93][94][95][96][97][98][99][100][101][102][103][104][105][106][107][108][109][110][111][112][113][114][115][116][117][110][118][119][120][121][122][123][124][125][126][127][128][129][130][131][132][133][134][135][136][137][138][139][110][140][141][142][143][144][145][5][146][147][5][148][149][150][151][152][153][154][155][156][157][158][159][160][161][162][5][163][164][165][166][5][167][168][169][170][171][172][173][174][175][176][177][178][179][5][180][181][182][183][184][185][186][187][188][189][5][190][191][110][192][193][194][5][195][196][197][198][199][200][201][202][203][204][205][206][207][208][209][110][210][211][5][212][213][214][215][216][217][110][218][219][220][221][222][223][224][225][226][227][228][229][230][231][232][233][234][235][110][236][237][238][239][240][241][242][243][244][245][246][247][248][249][250][251][252][253][254][255][256][257][258][259][260][261][262][263][264][265][266][267][5][268][110][269][270][271][272][273][274][275][276][277][278][279][280][281][282][283][284][285][286][287][288][289][290][291][292][293][294][295][296][5][297][298][299][5][300][301][302][303][304][305][306][307][308][309][5][310][311][312][313][314][315][316][317][318][319][320][321][322][5][323][324][325][326][327][328][329][330][110][331][332][5]
