Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Amanda Izeli Portilho | + 7211 word(s) | 7211 | 2022-03-14 04:50:39 | | | |
| 2 | Conner Chen | -5307 word(s) | 1904 | 2022-03-24 01:18:01 | | | | |
| 3 | Conner Chen | Meta information modification | 1904 | 2022-03-24 01:18:22 | | | | |
| 4 | Conner Chen | Meta information modification | 1904 | 2022-03-24 02:11:16 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Portilho, A. Enzyme-Linked Immunosorbent Assay. Encyclopedia. Available online: https://encyclopedia.pub/entry/20935 (accessed on 08 February 2026).
Portilho A. Enzyme-Linked Immunosorbent Assay. Encyclopedia. Available at: https://encyclopedia.pub/entry/20935. Accessed February 08, 2026.
Portilho, Amanda. "Enzyme-Linked Immunosorbent Assay" Encyclopedia, https://encyclopedia.pub/entry/20935 (accessed February 08, 2026).
Portilho, A. (2022, March 23). Enzyme-Linked Immunosorbent Assay. In Encyclopedia. https://encyclopedia.pub/entry/20935
Portilho, Amanda. "Enzyme-Linked Immunosorbent Assay." Encyclopedia. Web. 23 March, 2022.
Copy Citation
The Enzyme-Linked Immunosorbent Assay is a versatile technique, which can be used for several applications. It has enormously contributed to the study of infectious diseases.
ELISA
immune response
antibody
cytokine
SARS-CoV-2
1. Introduction
Enzyme-Linked Immunosorbent Assay (ELISA) is a widely used method, most of all, because of its flexibility: in-house assays can be performed or commercial kits can be used; it is possible to analyze several samples because of its high throughput capacity and lots of different analytes can be studied. Therefore, as long as it is standardized, the test has multiple applications [1].
When it comes to the immune response in the context of infectious diseases, ELISA is a technique that can provide a variety of data. For example, it supports diagnosis detecting microbial antigens in a sample; verifies prior exposure to the pathogen detecting antibodies; underpins epidemiologic monitoring with serologic surveys; and aids immune intervention studies, through lots of applications (e.g., supporting vaccine studies or filtering monoclonal antibodies) [2][3][4].
2. Detection of Immune Response to SARS-CoV-2
Several tests can be used to investigate the immune response. Choosing the more adequate assay should consider laboratory facilities, regarding both infrastructure and professional qualifications, as well as the question to be answered. Understanding the advantages and limitations of a serologic technique is the key not just to making the right choice, but also to address an adequate analysis [5][6][7].
3. Enzyme-Linked Immunosorbent and Other Immunoassays
Several immunoassays are used to study the immune response after vaccination or natural infection. In the case of ELISA, it allows high throughput flows, supported by automatic washers and readers. Usually, it is performed in 96-wells microplates. The first manuscripts proposing its use in SARS-CoV-2 studies consisted of protocols standardized in house [8][9]. The interesting point of such protocols is the possibility of re-standardizing it in different laboratories. However, commercial kits are also available, addressing the needs of clinical laboratories [10].
Chemiluminescence immunoassays allow complete automation and several kits are available now. However, a study found good correlation between ELISAs protocols and automated immunoassays, using both Nucleocapsid and Spike proteins as coating antigens [11].
ELISA can also be applied for multiplex analysis, using microarrays where different antigens are coated. Two examples are ViraChip® (ViraMed Biotech AG, Planegg, Niemcy) and multiSero assays, which use, respectively, subunits 1 (S1) and 2 (S2) of Spike (S) and Nucleocapsid (N) and S, N and Receptor-binding domain (RBD) of SARS-CoV-2. Reports of these tests reach 95 (multiSero) and 100% specificity (ViraChip®, ViraMed Biotech AG, Planegg, Niemcy) [12][13][14][15]. Thus, the results obtained distinguish the antigens triggering the humoral response, which supports immunologic and vaccine studies.
Usually, ELISA or chemiluminescence assays are more sensitive than immunocromatography [15][16]. However, a study found that the sensitivity of lateral flow immunoassays ranged from ≥92.1 to 100% when samples collected 14 days after the infection were tested [17]. It must be highlighted that sensitivity of immunoassays is related to sampling time. It might decrease when samples collected after several months are used, which seems to be a feature of SARS-CoV-2 immune response [18].
All in all, ELISA, as well as other techniques adapted from it, are capable of detecting the presence of SARS-CoV-2 antigens or antibodies in the sample tested. All these platforms are easy to perform and can provide results within hours. Hence, they can be used for diagnostics or research purposes [1][19]. Also, the methodologies can be adapted to describe other features of the immune response, as cytokine release, which helps to elucidate SARS-CoV-2 immune response. It is important to note that the question to be answered should guide both choice and standardization of the assay [2][5].
4. SARS-CoV-2 Antigens and Antibodies
The main antigens studied in SARS-CoV-2 humoral response are structural proteins Spike (S) and Nucleocapsid (N). It is observed that S protein can be detected entirely, or by means of its subunits 1 (S1) and 2 (S2). Another antigen related to it is the Receptor-binding domain (RBD), which lies at S1 [20].
Before discussing the antigens and immunoglobulin (Ig) classes, it is relevant to highlight that, in general, the detection of antibodies is a limited tool for diagnosis. In some cases, the detection of IgM or IgG may point out acute or chronic infection, respectively [21]. However, the pathophysiology of the disease should be well-known, so the results will not be masked by vaccine response or immunologic window. This is why detecting the pathogen itself, its nucleic acid, or antigens usually are considered a gold standard [22][23]. Concerning SARS-CoV-2, the gold standard for diagnosis is the detection of viral RNA by RTq-PCR. Considering the unprecedented number of cases, the use of antigen-detection was a strategy to support COVID-19 diagnosis, proving the presence of viral antigens in nasopharyngeal samples [23][24].
The abundance of S and N antigens in the course of SARS-CoV-2 infection substantiates its use in immunoassays, regardless of the aim being to detect the antigen or the antibodies [25]. It must be noted that there is a difference in the positivity dynamics of each antigen. Anti-N antibodies become positive, on average, two days earlier than anti-S antibodies, but they have a shorter half-life. In fact, anti-S antibodies are detectable for longer periods. Thus, it might explain decreased sensitivity of N-based assays when samples collected at longer moments are tested [26].
Antigen-detection assays are often based on lateral-flow immunoassays using nasopharyngeal samples, where the viral load is concentrated. The use of two antigens increases test sensitivity [23]. In fact, testing nasopharyngeal samples in an N-chemiluminescence enzymatic immunoassay provided satisfactory concordance with gold-standard RT-qPCR [27]. To achieve high sensitivity and specificity, antigen-detection tests should be conducted up to seven days after infection [28]. However, a fluorescent ELISA to detect the Nucleocapsid antigen in serum was developed, as well as an ELISA to capture the Spike protein [7][29]. To improve clinical-laboratory testing, automated assays to detect SARS-CoV-2 antigens are available as well, and correlated well with RT-qPCR results. In that case, specificity may reach 100%, while sensitivity levels are related to the positivity threshold established [30].
Considering antibody-detection assays, it should be understood that using S and N proteins might increase the sensitivity, providing more epitopes for the antibodies to bind. On the other hand, it can increase the cross-reactivity. Thus, subunit antigens, such as RBD, have been used to assess SARS-CoV-2 immune response in a more specific way [25]. This antigen is undeniably important, since it mediates the interaction between the virus and the Angiotensin-converting enzyme 2 (ACE-2), used as receptor. However, there are studies reporting neutralizing antibodies directed to the N-terminal domain (NTD) of S1 subunit, highlighting that this is another relevant antigen to be studied [31][32].
Envelope (E) and Membrane (M) are other structural proteins but, due to their small molecular size, usually they do not provide a robust humoral response [20]. However, recent studies provide new data about it. Anti-Membrane antibodies were detected in COVID-19 patients up to 12 months after infection and Envelope-IgM has shown a less accentuated decrease than S1-IgM [33]. Thus, tests detecting Ig against different antigens could be employed to distinguish natural and vaccine-induced immune responses, as long as the subjects were vaccinated with subunit vaccines.
Considering Ig classes, the curve of anti-SARS-CoV-2 antibodies differs from most infections, making it difficult to predict acute or chronic/memory immune responses. Usually, IgM antibodies increase during the first two weeks of infection; hence, its “acute marker” characteristic was coined. Afterwards, IgM antibodies decay while IgG titers increase [21]. In COVID-19, the first reports suggested the same pattern, but more recent studies point to a different one, where IgM and IgG antibodies increase concomitantly since the beginning of infection, being detected as early as three days following infection [34][35]. Even more curious is the fact that IgA would be a better marker of acute infection than IgM, increasing in the first days following infection and maintaining its titers for up to 30 days, whereas IgM would decay after 15 days. Also, combined detection of IgA/IgG would confer more sensitive results than IgM/IgG. Another way to increase the sensitivity of immunoassays is to use anti-Ig detection antibodies, which will reveal the presence of the whole antibody content of the sample, regardless of the Ig class [36].
IgG is related to immunologic memory and, in some cases, it can be detected years after the infection or vaccination [21]. However, serologic studies suggest that anti-SARS-CoV-2 IgG would decrease six to eight months after infection [37][38]. Although the vaccination campaigns are recent and studies to elucidate the immunologic memory conferred by them are still ongoing, there are reports showing persistence of antibodies up to six months after vaccination [39][40]. It is worth emphasizing that seropositivity for anti-SARS-CoV-2 IgM and IgG occurs about the same time following infection.
Figure 1 represents the structural antigens used in SARS-CoV-2 immunoassays and the antigen or antibody detection curve expected in SARS-CoV-2 immune response.
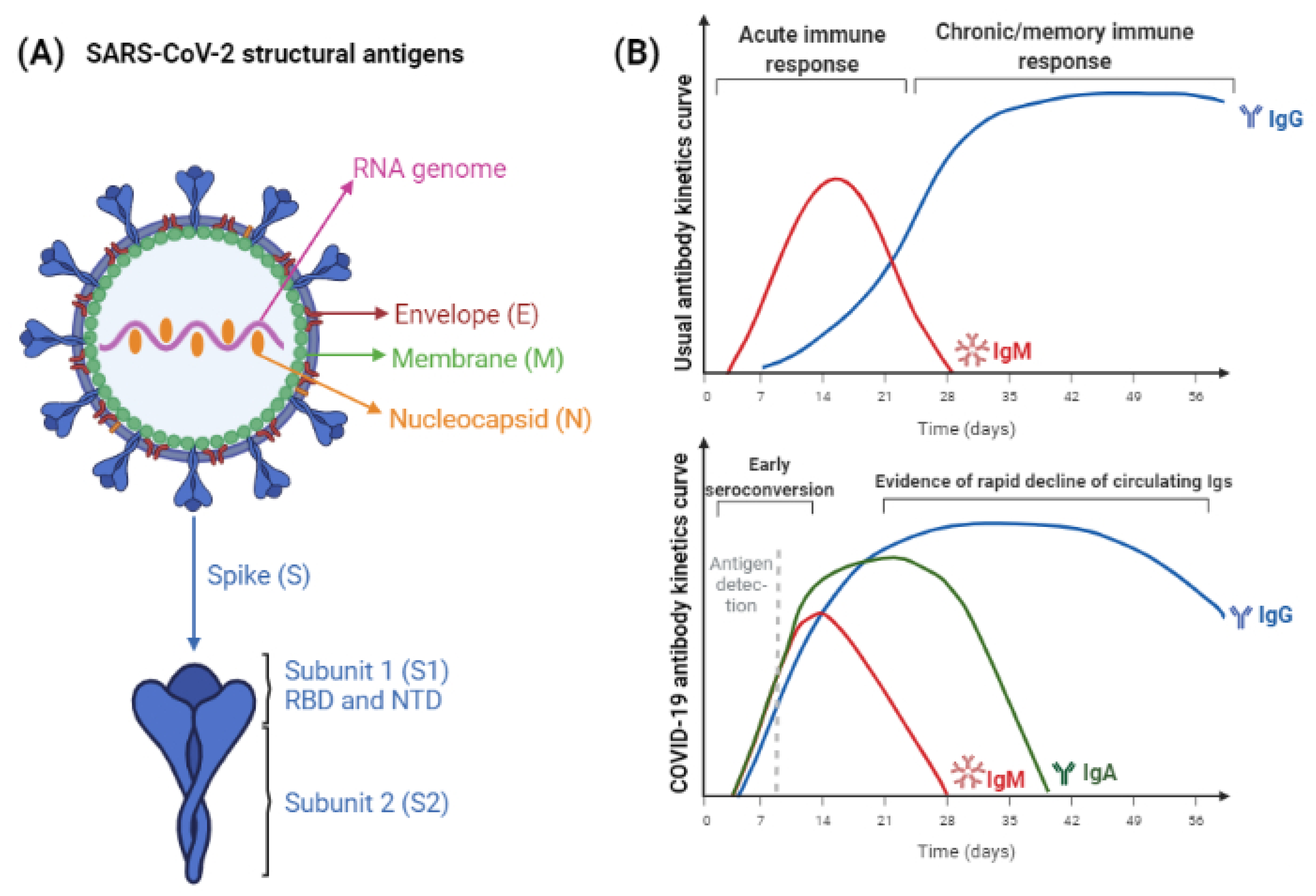
Figure 1. (A) SARS-CoV-2 structural antigens, used for immunoassays. Nucleocapsid and Spike proteins are the most abundant antigens, therefore, they are used for improved sensitivity. Antigen-subunits, as S1 and S2, or RBD and NTD, can provide more specific results. (B) The expected IgM/IgG curves following exposure to antigen through natural infection or vaccination. The maturation of immune response usually leads to initial IgM induction and, while titers of this antibody class decrease, IgG titers increase and persist for longer periods, which varies according to the pathogen considered. Below it, kinetics of SARS-CoV-2 antigens and antibodies detection is represented. Differently from the expected, titers of IgA, IgM, and IgG antibodies increase in parallel. The time to achieve seronegativity and the particularities of vaccine-induced humoral response are still being elucidated (figure created with BioRender).
The sole presence of antibodies following natural infection should be cautiously analyzed, especially when the patient is individually considered, because it does not prove an antibody functionality, predicting protection [2]. So far, high antibodies quantities were observed in severe COVID-19 cases [41][42].
For some infectious diseases, the specificity of the immune response can predict better or worse prognostic. Some studies following COVID-19 patients suggested a worst prognosis when a Nucleocapsid was observed instead of a Spike-biased immune response. However, others could not observe any difference between the antigens recognized and the severity of the disease [41][42]. Similarly, the impact of the kinetics of humoral response was related to a better or worst clinical condition. Hashem et al. [43] reported that an early seroconversion correlated with fatality, but Lucas et al. [44] observed a delayed antibody response in deceased patients. Meanwhile, the study of Van Elslande et al. [17] did not indicate significant differences in time to critical and non-critical patients to seroconvert. Disagreeing results suggest that further studies are necessary to elucidate these aspects and support the interpretation of ELISA results.
Secretory IgA, locally produced at mucosal sites, are chief to control mucosal infections, but the role of seric IgA has been up for debate. It may help control the inflammation, but authors disagree about its capacity to activate the complement system and opsonize pathogens [45][46][47]. Regarding COVID-19, there is evidence that seric IgA acts on early virus neutralization [48]. Fedele et al. [49] observed that early IgA response was seen in patients with less severe COVID-19, but Portilho et al. [42] could not relate increased IgA response to mild patients response. Despite the disagreeing results of different studies, the detection of seric IgA could reflect secretory IgA. There is evidence that naïve B cells activated in the mucosa can be home to the marginal zone of the spleen, so IgA triggered against mucosal pathogens could be released in the bloodstream and contribute to control the infection [50][51].
Finally, even though antibody-detection assays are limited to diagnosing the current infection, these technologies are still relevant, especially to select plasma donors, vaccine studies, and epidemiological monitoring [2].
References
- Crowther, J.R. The ELISA Guidebook, 2nd ed.; Humana Press Inc.: Vienna, Austria, 2009; ISBN 978-1-60327-253-7.
- West, R.; Kobokovich, A.; Connell, N.; Gronvall, G.K. COVID-19 Antibody Tests: A Valuable Public Health Tool with Limited Relevance to Individuals. Trends Microbiol. 2021, 29, 214–223.
- Maggi, E.; Canonica, G.W.; Moretta, L. COVID-19: Unanswered questions on immune response and pathogenesis. J. Allergy Clin. Immunol. 2020, 146, 18–22.
- Assadiasl, S.; Fatahi, Y.; Zavvar, M.; Nicknam, M.H. COVID-19: Significance of antibodies. Hum. Antibodies 2020, 28, 287–297.
- Ong, D.S.Y.; Fragkou, P.C.; Schweitzer, V.A.; Chemaly, R.F.; Moschopoulos, C.D.; Skevaki, C.; European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group for Respiratory Viruses (ESGREV). How to interpret and use COVID-19 serology and immunology tests. Clin. Microbiol. Infect. 2021, 27, 981–986.
- Hamilton, R.G. The clinical immunology laboratory of the future. Clin. Chem. 1994, 40, 2186–2192.
- Liu, G.; Rusling, J.F. COVID-19 Antibody Tests and Their Limitations. ACS Sensors 2021, 6, 593–612.
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.O.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26, 1033–1036.
- Moura, A.D.; Costa, H.H.M.; Correa, V.A.; Lima, A.K.S.; Lindoso, J.A.L.; De Gaspari, E.; Hong, M.A.; Cunha-Junior, J.P.; Prudencio, C.R. Assessment of avidity related to IgG subclasses in SARS-CoV-2 Brazilian infected patients. Sci. Rep. 2021, 11, 17642.
- Allinson, J.L. Automated immunoassay equipment platforms for analytical support of pharmaceutical and biopharmaceutical development. Bioanalysis 2011, 3, 2803–2816.
- Van Elslande, J.; Decru, B.; Jonckheere, S.; Van Wijngaerden, E.; Houben, E.; Vandecandelaere, P.; Indevuyst, C.; Depypere, M.; Desmet, S.; André, E.; et al. Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin. Microbiol. Infect. 2020, 26, 1557.e1–1557.e7.
- Byrum, J.R.; Waltari, E.; Janson, O.; Guo, S.-M.; Folkesson, J.; Chhun, B.B.; Vinden, J.; Ivanov, I.E.; Forst, M.L.; Li, H.; et al. multiSero: Open multiplex-ELISA platform for analyzing antibody responses to SARS-CoV-2 infection. MedRxiv 2021.
- Krüttgen, A.; Cornelissen, C.G.; Dreher, M.; Hornef, M.; Imöl, M.; Kleines, M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J. Clin. Virol. 2020, 128, 104394.
- Olbrich, L.; Castelletti, N.; Schälte, Y.; Garí, M.; Pütz, P.; Bakuli, A.; Pritsch, M.; Kroidl, I.; Saathoff, E.; Guggenbuehl Noller, J.M.; et al. Head-to-head evaluation of seven different seroassays including direct viral neutralisation in a representative cohort for SARS-CoV-2. J. Gen. Virol. 2021, 102, 001653.
- Bastos, L.M.; Tavaziva, G.; Abidi, S.K.; Campbell, J.R.; Haraoui, L.P.; Johnston, J.C.; Lan, Z.; Law, S.; MacLean, E.; Trajman, A.; et al. Diagnostic accuracy of serological tests for covid-19: Systematic review and meta-analysis. BMJ 2020, 370, m2516.
- Saker, K.; Escuret, V.; Pitiot, V.; Massardier-Pilonchéry, M.; Paul, S.; Mokdad, B.; Langlois-Jaques, C.; Rabilloud, M.; Goncalves, D.; Fabien, N.; et al. Evaluation of commercial anti-SARS-CoV-2 antibody assays and comparison of standardized titers in vaccinated health care workers. J. Clin. Microbiol. 2021, 60, e01746-21.
- Van Elslande, J.; Houben, E.; Depypere, M.; Brackenier, A.; Desmet, S.; André, E.; van Ranst, M.; Lagrou, K.; Vermeersch, P. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin. Microbiol. Infect. 2020, 26, 1082–1087.
- Perez-Saez, J.; Zaballa, M.E.; Yerly, S.; Andrey, D.O.; Meyer, B.; Eckerlle, I.; Balavoine, J.-F.; Chappuis, F.; Pittet, D.; Trono, D.; et al. Persistence of anti-SARS-CoV-2 antibodies: Immunoassay heterogeneity and implications for serosurveillance. Clin. Microbiol. Infect. 2021, 27, 1695.e7–1695.e12.
- Dowlatshahi, S.; Shabani, E.; Abdekhodaie, M.J. Serological assays and host antibody detection in coronavirus-related disease diagnosis. Arch. Virol. 2021, 166, 715–731.
- Dai, L.; Gao, G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021, 21, 73–82.
- Mesquita Júnior, D.; Araújo, J.A.P.; Catelan, T.T.T.; Souza, A.W.S.; Cruvinel, W.M.; Andrade, L.E.C.; Silva, N.P. Immune System Part II—Basis of the immunological response mediated by T and B lymphocytes. Brazilian J. Rheumatol. 2010, 50, 552–580.
- Cancrini, G.; Iori, A. Traditional and innovative diagnostic tools: When and why should be applied. Parassitologia 2004, 46, 173–176.
- Ilkhani, H.; Hedayat, N.; Farhad, S. Novel approaches for rapid detection of COVID-19 during the pandemic: A review. Anal. Biochem. 2021, 634, 114362.
- Matsuda, E.M.; de Campos, I.B.; de Oliveira, I.P.; Colpas, D.R.; dos Santos Carmo, A.M.; Brígido, L.F.M. Field evaluation of COVID-19 antigen tests versus RNA based detection: Potential lower sensitivity compensated by immediate results, technical simplicity, and low cost. J. Med. Virol. 2021, 93, 4405–4410.
- Khan, J.; Al Asoom, L.I.; Khan, M.; Chakrabartty, I.; Dandoti, S.; Rudrapal, M.; Zothantluanga, J.H. Evolution of RNA viruses from SARS to SARS-CoV-2 and diagnostic techniques for COVID-19: A review. J. Basic Appl. Sci. 2021, 10, 60.
- Lumley, S.F.; Wei, J.; O’Donnel, D.; Stoesser, N.E.; Matthews, P.C.; Howarth, A.; Hatch, S.B.; Marsden, B.D.; Cox, S.; James, T.; et al. The Duration, Dynamics, and Determinants of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Antibody Responses in Individual Healthcare Workers. Clin. Infect. Dis. 2021, 73, e699–e709.
- Villalta, D.; Moratto, A.; Salgarolo, V.; Da Re, M.; Giacomello, R.; Malipiero, G. New-Generation Quantitative Immunoassays for SARS-CoV-2 Antibody Detection: Need for Harmonization. Ann. Lab. Med. 2022, 42, 113–116.
- Abusrewill, Z.; Alhudiri, I.M.; Kaal, H.H.; El Meshri, S.E.; Ebrahim, F.O.; Dalyoum, T.; Efrefer, A.A.; Ibrahim, K.; Elfghi, M.B.; Abusrewill, S.; et al. Time scale performance of rapid antigen testing for SARS-CoV-2: Evaluation of 10 rapid antigen assays. J. Med. Virol. 2021, 93, 6512–6518.
- Girt, G.C.; Lakshminarayanan, A.; Huo, J.; Dormon, J.; Norman, C.; Afrough, B.; Harding, A.; James, W.; Owens, R.J.; Naismith, J.H. The use of nanobodies in a sensitive ELISA test for SARS-CoV-2 Spike 1 protein. R. Soc. Open Sci. 2021, 8, 211016.
- Van der Moeren, N.; Zwart, V.F.; Goderski, G.; Rijkers, G.T.; van den Bijllaardt, W.; Veenemans, J.; Kluytmans, J.A.J.W.; Pas, S.D.; Meijer, A.; Verweij, J.J.; et al. Performance of the Diasorin SARS-CoV-2 antigen detection assay on the LIAISON XL. J. Clin. Virol. 2021, 141, 104909.
- Amanat, F.; Thapa, M.; Lei, T.; Ahmed, S.M.S.; Adelsberg, D.C.; Carreño, J.M.; Strohmeier, S.; Schmitz, A.J.; Zafar, S.; Zhou, J.Q.; et al. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell 2021, 184, 3936–3948.
- Krut, V.G.; Astrakhantseva, I.V.; Chuvpilo, S.A.; Efimov, G.A.; Ambaryan, S.G.; Drutskaya, M.S.; Nedospasov, S.A. Antibodies to the N-Terminal Domain of Angiotensin-Converting Enzyme (ACE2) That Block Its Interaction with SARS-CoV-2 S Protein. Dokl. Biochem. Biophys. 2021, 1–4.
- Amjadi, M.F.; Adyniec, R.R.; Gupta, S.; Bashar, S.J.; Mergaert, A.M.; Braun, K.M.; Moreno, G.K.; O’Connor, D.H.; Friedrich, T.C.; Safdar, N.; et al. Anti-membrane and anti-spike antibodies are long-lasting and together discriminate between past COVID-19 infection and vaccination. medRxiv 2021.
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody Responses to SARS-CoV-2 in Patients with Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2027–2034.
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506.
- Ma, H.; Zeng, W.; He, H.; Zhao, D.; Jiang, D.; Zhou, P.; Cheng, L.; Li, Y.; Ma, X.; Jin, T. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020, 17, 773–775.
- Liu, C.; Yu, X.; Gao, C.; Zhang, L.; Zhai, H.; Hu, Y.; Liu, E.; Wang, Q.; Gao, Y.; Wei, D.; et al. Characterization of antibody responses to SARS-CoV-2 in convalescent COVID-19 patients. J. Med. Virol. 2021, 93, 2227–2233.
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371, eabf4063.
- Swadzba, J.; Anyszek, T.; Panek, A.; Martin, E. Anti-Spike SARS-CoV-2 IgG Assessment with a Commercial Assay during a 4-Month Course after COVID-19 Vaccination. Vaccines 2021, 9, 1367.
- Ciabattini, A.; Pastore, G.; Fiorino, F.; Polvere, J.; Lucchesi, S.; Pettini, E.; Stefano, A.; Rancan, I.; Durante, M.; Miscia, M.; et al. Evidence of SARS-CoV-2-Specific Memory B Cells Six Months After Vaccination With the BNT162b2 mRNA Vaccine. Front. Immunol. 2021, 12, 740708.
- Atyeo, C.; Fischinger, S.; Zohar, T.; Slein, M.D.; Burke, J.; Loos, C.; McCulloch, D.J.; Newman, K.L.; Wolf, C.; Yu, J.; et al. Distinct Early Serological Signatures Track with SARS-CoV-2 Survival. Immunity 2020, 53, 524–532.E4.
- Portilho, A.I.; Silva, V.O.; Ahagon, C.M.; Matsuda, E.M.; de Oliveira, E.L.; da Silveira, E.P.R.; Lima, A.K.S.; Lindoso, J.A.L.; Campos, I.B.; Hong, M.A.; et al. Humoral response to spike S1 and S2 and nucleocapsid proteins on microarray after SARS-CoV-2 infection. J. Med. Virol. 2021, 94, 178–185.
- Hashem, A.; Algaissi, A.; Almahboub, S.; Alfaleh, M.; Abujamel, T.; Alamri, S.; Alluhaybi, K.; Hobani, H.; AlHarbi, R.; Alsulaiman, R.; et al. Early Humoral Response Correlates with Disease Severity and Outcomes in COVID-19 Patients. Viruses 2020, 12, 1390.
- Silva, J.; Lucas, C.; Sundaram, M.; Israelow, B.; Wong, P.; Klein, J.; Lu, P.; Venkataraman, A.; Liu, F.; Mao, T.; et al. Saliva viral load is a dynamic unifying correlate of COVID-19 severity and mortality. medRxiv 2021.
- Mestecky, J.; Mcghee, J.R. Immunoglobulin A (IgA): Molecular and Cellular Interactions Involved in IgA Biosynthesis and Immune Response. Adv. Immunol. 1987, 40, 153–245.
- Kerr, M.A. The structure and function of human IgA. Biochem. J. 1990, 271, 285–296.
- Hiemstra, P.S.; Gorter, A.; Stuurman, M.E.; van Es, L.A.; Daha, M.R. Activation of the alternative pathway of complement by human serum IgA. Adv. Exp. Med. Biol. 1987, 17, 321–326.
- Sterlin, D.; Mathian, A.; Miyara, M.; Mohr, A.; Anna, F.; Claër, L.; Quentric, P.; Fadlallah, J.; Ghillani, P.; Gunn, C.; et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021, 13, eabd2223.
- Fedele, G.; Russo, G.; Schiavoni, I.; Leone, P.; Olivetta, E.; Perri, E.; Zingaropoli, M.A.; Ciardi, M.R.; Pasculli, P.; Mastroianni, C.M.; et al. Early IgG / IgA response in hospitalized COVID-19 patients is associated with a less severe disease. Diagn. Microbiol. Infect. Dis. 2022, 102, 115586.
- Vossenkämper, A.; Blair, P.A.; Safinia, N.; Fraser, L.D.; Das, L.; Sanders, T.J.; Stagg, A.J.; Sanderson, J.D.; Taylor, K.; Chang, F.; et al. A role for gut-associated lymphoid tissue in shaping the human B cell repertoire. J. Exp. Med. 2013, 210, 1665–1674.
- Victora, G.D.; Nussenzweig, M.C. Germinal Centers. Ann. Rev. Immunol. 2012, 30, 429–457.
More
Information
Subjects:
Immunology; Virology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Entry Collection:
COVID-19
Revisions:
4 times
(View History)
Update Date:
24 Mar 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




