
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Geovani Lopez-Ortiz | + 2488 word(s) | 2488 | 2022-01-26 09:30:47 | | | |
| 2 | Lindsay Dong | Meta information modification | 2488 | 2022-02-25 03:49:49 | | |
Video Upload Options
From the start of the COVID-19 pandemic, new SARS-CoV-2 variants have emerged that potentially affect transmissibility, severity, and immune evasion in infected individuals. Conclusions: SARS-CoV-2 variants can potentially have an impact on clinical outcomes.
1. Introduction
2. SARS-CoV-2 Variants and Clinical Outcomes
2.1. SARS-CoV-2 Variants
| Changes | Location | Sources |
|---|---|---|
|
Spike protein (S) | [6][16][17][18][19][20][21][22][24][25][26][27][28][29][30][33][34][35][36][37][38] |
|
Nucleocapsid phosphoprotein (N protein) | [39][18][22][24][27][29][30][37][38] |
|
ORF1a | [39][21][26] |
|
NSP1 | [38] |
|
NSP2 | [24][27][33][38] |
|
NSP3 | [39][18][27][33][35][36][38] |
|
NSP4 | [18][27][29][33][35][38] |
|
NSP7 | [38] |
|
NSP8 | [27][38] |
|
NSP9 | [27] |
|
NSP12 | [39][22][35] |
|
NSP13 | [27][33][35][38] |
|
NSP14 | [33][35] |
|
NSP15 | [27] |
|
NSP16 | [27] |
|
ORF3a | [39][18][22][24][27][29][30][33][36][37][38][40] |
|
ORF6 | [18][37] |
|
ORF7b | [37] |
|
ORF8 | [17][18][29][30][33][35][40][41] |
|
RdRp | [39][17][18][19][21][24][27][29][30][33][36][37][38][42] |
|
3C-like protease | [38] |
2.2. SARS-CoV-2 Variants and Clinical Outcomes
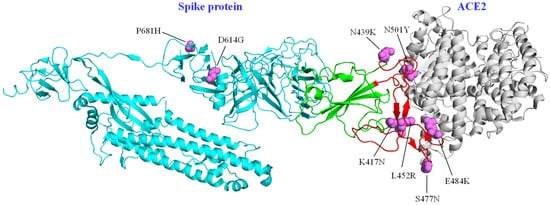
Prior to the reporting of Variants of Interest (VOI) and Variants of Concern (VOC), changes in the SARS-CoV-2 sequence that could have an impact on clinical outcomes had been determined [6]. The D614G variant in the spike protein was initially considered to be related to a higher rate of hospitalizations and moderate to severe clinical outcomes [6][17]; however, analyses in different cohorts showed no relationship with disease severity; this change increases the adaptability of the virus in human populations, without necessarily causing more severe disease [19][37]. The same scenario was visualized for the N439K variant in the spike protein, which was also not found to have a direct effect on clinical outcomes, compared to the original virus. However, it was reported that this substitution had emerged in different clades independently and that it increased affinity for ACE2 and resistance against various neutralizing monoclonal antibodies [25].
2.3. Rise and Spread of Variants of Concern
2.4. Other Variants Related with Clinical Outcomes
3. Conclusions
References
- Vijay, N.; Weissensteiner, M.; Burri, R.; Kawakami, T.; Ellegren, H.; Wolf, J.B.W. Genomewide patterns of variation in genetic diversity are shared among populations, species and higher-order taxa. Mol. Ecol. 2017, 26, 4284–4295.
- Franzo, G.; Drigo, M.; Legnardi, M.; Grassi, L.; Pasotto, D.; Menandro, M.; Cecchinato, M.; Tucciarone, C. Bovine Coronavirus: Variability, Evolution, and Dispersal Patterns of a No Longer Neglected Betacoronavirus. Viruses 2020, 12, 1285.
- Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a Novel Swine-Origin Influenza A (H1N1) Virus in Humans. N. Engl. J. Med. 2009, 360, 2605–2615.
- Torres, M.; de Mendonça, M.L.; Rodrigues, C.D.D.S.; Fonseca, V.; Ribeiro, M.; Brandão, A.; da Cunha, R.V.; Dias, A.; Boas, L.S.V.; Felix, A.; et al. Dengue Virus Serotype 2 Intrahost Diversity in Patients with Different Clinical Outcomes. Viruses 2021, 13, 349.
- Tsetsarkin, K.A.; VanLandingham, D.L.; McGee, C.E.; Higgs, S. A Single Mutation in Chikungunya Virus Affects Vector Specificity and Epidemic Potential. PLoS Pathog. 2007, 3, e201.
- Eaaswarkhanth, M.; Al Madhoun, A.; Al-Mulla, F. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int. J. Infect. Dis. 2020, 96, 459–460.
- Chadha, J.; Khullar, L.; Mittal, N. Facing the wrath of enigmatic mutations: A review on the emergence of severe acute respiratory syndrome coronavirus 2 variants amid coronavirus disease-19 pandemic. Environ. Microbiol. 2021.
- Mansbach, R.A.; Chakraborty, S.; Nguyen, K.; Montefiori, D.C.; Korber, B.; Gnanakaran, S. The SARS-CoV-2 Spike variant D614G favors an open conformational state. Sci. Adv. 2021, 7, eabf3671.
- Almubaid, Z.; Al-Mubaid, H. Analysis and comparison of genetic variants and mutations of the novel coronavirus SARS-CoV-2. Gene Rep. 2021, 23, 101064.
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424.
- Stirrup, O.; Boshier, F.; Venturini, C.; Guerra-Assunção, J.A.; Alcolea-Medina, A.; Beckett, A.; Charalampous, T.; Filipe, A.D.S.; Glaysher, S.; Khan, T.; et al. SARS-CoV-2 lineage B.1.1.7 is associated with greater disease severity among hospitalised women but not men: Multicentre cohort study. BMJ Open Respir. Res. 2021, 8, e001029.
- Anastassopoulou, C.; Gkizarioti, Z.; Patrinos, G.P.; Tsakris, A. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum. Genom. 2020, 14, 1–8.
- Patone, M.; Thomas, K.; Hatch, R.; Tan, P.S.; Coupland, C.; Liao, W.; Mouncey, P.; Harrison, D.; Rowan, K.; Horby, P.; et al. Mortality and critical care unit admission associated with the SARS-CoV-2 lineage B.1.1.7 in England: An observational cohort study. Lancet Infect. Dis. 2021, 21, 1518–1528.
- Lin, L.; Liu, Y.; Tang, X.; He, D. The Disease Severity and Clinical Outcomes of the SARS-CoV-2 Variants of Concern. Front. Public Health 2021, 9, 775224.
- Courjon, J.; Contenti, J.; Demonchy, E.; Levraut, J.; Barbry, P.; Rios, G.; Dellamonica, J.; Chirio, D.; Bonnefoy, C.; Giordanengo, V.; et al. COVID-19 patients age, comorbidity profiles and clinical presentation related to the SARS-CoV-2 UK-variant spread in the Southeast of France. Sci. Rep. 2021, 11, 18456.
- Jin, X.; Lian, J.S.; Hu, J.H.; Gao, J.; Zheng, L.; Zhang, Y.M.; Hao, S.R.; Jia, H.Y.; Cai, H.; Zhang, X.L.; et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 2020, 69, 1002–1009.
- Elizondo, V.; Harkins, G.W.; Mabvakure, B.; Smidt, S.; Zappile, P.; Marier, C.; Maurano, M.T.; Perez, V.; Mazza, N.; Beloso, C.; et al. SARS-CoV-2 genomic characterization and clinical manifestation of the COVID-19 outbreak in Uruguay. Emerg. Microbes Infect. 2021, 10, 51–65.
- Nagy, Á.; Pongor, S.; Győrffy, B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int. J. Antimicrob. Agents 2021, 57, 106272.
- Isabel, S.; Graña-Miraglia, L.; Gutierrez, J.M.; Bundalovic-Torma, C.; Groves, H.E.; Isabel, M.R.; Eshaghi, A.; Patel, S.N.; Gubbay, J.B.; Poutanen, T.; et al. Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide. Sci. Rep. 2020, 10, 14031.
- Müller, N.F.; Wagner, C.; Frazar, C.D.; Roychoudhury, P.; Lee, J.; Moncla, L.H.; Pelle, B.; Richardson, M.; Ryke, E.; Xie, H.; et al. Viral genomes reveal patterns of the SARS-CoV-2 outbreak in Washington State. Sci. Transl. Med. 2021, 13, eabf0202.
- Al Khatib, H.A.; Benslimane, F.M.; Elbashir, I.E.; Coyle, P.V.; Al Maslamani, M.A.; Al-Khal, A.; Al Thani, A.A.; Yassine, H.M. Within-Host Diversity of SARS-CoV-2 in COVID-19 Patients with Variable Disease Severities. Front. Cell. Infect. Microbiol. 2020, 10, 575613.
- Fournier, P.-E.; Colson, P.; Levasseur, A.; Devaux, C.A.; Gautret, P.; Bedotto, M.; Delerce, J.; Brechard, L.; Pinault, L.; Lagier, J.-C.; et al. Emergence and outcomes of the SARS-CoV-2 ‘Marseille-4’ variant. Int. J. Infect. Dis. 2021, 106, 228–236.
- Hoang, V.-T.; Colson, P.; Levasseur, A.; Delerce, J.; Lagier, J.-C.; Parola, P.; Million, M.; Fournier, P.-E.; Raoult, D.; Gautret, P. Clinical outcomes in patients infected with different SARS-CoV-2 variants at one hospital during three phases of the COVID-19 epidemic in Marseille, France. Infect. Genet. Evol. 2021, 95, 105092.
- Morris, C.P.; Luo, C.H.; Amadi, A.; Schwartz, M.; Gallagher, N.; Ray, S.C.; Pekosz, A.; Mostafa, H.H. An Update on Severe Acute Respiratory Syndrome Coronavirus 2 Diversity in the US National Capital Region: Evolution of Novel and Variants of Concern. Clin. Infect. Dis. 2021, ciab636.
- Thomson, E.C.; Rosen, L.E.; Shepherd, J.G.; Spreafico, R.; Filipe, A.D.S.; Wojcechowskyj, J.A.; Davis, C.; Piccoli, L.; Pascall, D.J.; Dillen, J.; et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 2021, 184, 1171–1187.e20.
- Fillâtre, P.; Dufour, M.-J.; Behillil, S.; Vatan, R.; Reusse, P.; Gabellec, A.; Velmans, N.; Montagne, C.; Du Coudret, S.G.; Droumaguet, E.; et al. A new SARS-CoV-2 variant with high lethality poorly detected by RT-PCR on nasopharyngeal samples: An observational study. Clin. Microbiol. Infect. 2021, 28, 298.e9–298.e15.
- Gunadi; Wibawa, H.; Hakim, M.S.; Marcellus; Trisnawati, I.; El Khair, R.; Triasih, R.; Irene; Afiahayati; Iskandar, K.; et al. Molecular epidemiology of SARS-CoV-2 isolated from COVID-19 family clusters. BMC Med. Genom. 2021, 14, 144.
- Zhao, S.; Lou, J.; Chong, M.; Cao, L.; Zheng, H.; Chen, Z.; Chan, R.; Zee, B.; Chan, P.; Wang, M. Inferring the Association between the Risk of COVID-19 Case Fatality and N501Y Substitution in SARS-CoV-2. Viruses 2021, 13, 638.
- Nagy, Á.; Ligeti, B.; Szebeni, J.; Pongor, S.; Győrffy, B. COVIDOUTCOME—estimating COVID severity based on mutation signatures in the SARS-CoV-2 genome. Database 2021, 2021, baab020.
- Pang, X.; Li, P.; Zhang, L.; Que, L.; Dong, M.; Xie, B.; Wang, Q.; Wei, Y.; Xie, X.; Li, L.; et al. Emerging Severe Acute Respiratory Syndrome Coronavirus 2 Mutation Hotspots Associated with Clinical Outcomes and Transmission. Front. Microbiol. 2021, 12, 753823.
- Benton, D.J.; Wrobel, A.G.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588, 327–330.
- Benton, D.J.; Gamblin, S.J. SARS-CoV-2 Spike Glycoprotein with 2 ACE2 Bound. 2020. Available online: https://www.rcsb.org/structure/7a97 (accessed on 19 January 2022).
- Nakamichi, K.; Shen, J.Z.; Lee, C.S.; Lee, A.; Roberts, E.A.; Simonson, P.D.; Roychoudhury, P.; Andriesen, J.; Randhawa, A.K.; Mathias, P.C.; et al. Hospitalization and mortality associated with SARS-CoV-2 viral clades in COVID-19. Sci. Rep. 2021, 11, 1–11.
- Gunadi; Hakim, M.S.; Wibawa, H.; Marcellus; Trisnawati, I.; Supriyati, E.; Afiahayati; El Khair, R.; Iskandar, K.; Siswanto; et al. Association between prognostic factors and the outcomes of patients infected with SARS-CoV-2 harboring multiple spike protein mutations. Sci. Rep. 2021, 11, 21352.
- Cao, C.; He, L.; Tian, Y.; Qin, Y.; Sun, H.; Ding, W.; Gui, L.; Wu, P. Molecular epidemiology analysis of early variants of SARS-CoV-2 reveals the potential impact of mutations P504L and Y541C (NSP13) in the clinical COVID-19 outcomes. Infect. Genet. Evol. 2021, 92, 104831.
- Esper, F.P.; Cheng, Y.-W.; Adhikari, T.M.; Tu, Z.J.; Li, D.; Li, E.A.; Farkas, D.H.; Procop, G.W.; Ko, J.S.; Chan, T.A.; et al. Genomic Epidemiology of SARS-CoV-2 Infection During the Initial Pandemic Wave and Association with Disease Severity. JAMA Netw. Open 2021, 4, e217746.
- Siqueira, J.D.; Goes, L.R.; Alves, B.M.; de Carvalho, P.S.; Cicala, C.; Arthos, J.; Viola, J.P.B.; de Melo, A.C.; Soares, M. SARS-CoV-2 genomic analyses in cancer patients reveal elevated intrahost genetic diversity. Virus Evol. 2021, 7, veab013.
- Zekri, A.-R.N.; Mohanad, M.; Hafez, M.M.; Soliman, H.K.; Hassan, Z.K.; Abouelhoda, M.; Amer, K.E.; Seadawy, M.G.; Ahmed, O.S. Genome sequencing of SARS-CoV-2 in a cohort of Egyptian patients revealed mutation hotspots that are related to clinical outcomes. Biochim. Biophys. Acta 2021, 1867, 166154.
- Mehta, P.; Alle, S.; Chaturvedi, A.; Swaminathan, A.; Saifi, S.; Maurya, R.; Chattopadhyay, P.; Devi, P.; Chauhan, R.; Kanakan, A.; et al. Clinico-Genomic Analysis Reveals Mutations Associated with COVID-19 Disease Severity: Possible Modulation by RNA Structure. Pathogens 2021, 10, 1109.
- de Sousa, E.; Ligeiro, D.; Lérias, J.R.; Zhang, C.; Agrati, C.; Osman, M.; El-Kafrawy, S.A.; Azhar, E.I.; Ippolito, G.; Wang, F.-S.; et al. Mortality in COVID-19 disease patients: Correlating the association of major histocompatibility complex (MHC) with severe acute respiratory syndrome 2 (SARS-CoV-2) variants. Int. J. Infect. Dis. 2020, 98, 454–459.
- Young, B.E.; Wei, W.E.; Fong, S.-W.; Mak, T.-M.; Anderson, D.E.; Chan, Y.-H.; Pung, R.; Heng, C.S.; Ang, L.W.; Zheng, A.K.E.; et al. Association of SARS-CoV-2 clades with clinical, inflammatory and virologic outcomes: An observational study. EBioMedicine 2021, 66, 103319.
- Li, Z.; Li, Y.; Sun, R.; Li, S.; Chen, L.; Zhan, Y.; Xie, M.; Yang, J.; Wang, Y.; Zhu, A.; et al. Longitudinal virological changes and underlying pathogenesis in hospitalized COVID-19 patients in Guangzhou, China. Sci. China Life Sci. 2021, 64, 2129–2143.
- Ong, S.W.X.; Chiew, C.J.; Ang, L.W.; Mak, T.-M.; Cui, L.; Toh, M.P.H.; Lim, Y.D.; Lee, P.H.; Lee, T.H.; Chia, P.Y.; et al. Clinical and Virological Features of SARS-CoV-2 Variants of Concern: A Retrospective Cohort Study Comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). SSRN J. 2021, ciab721.
- Taylor, C.A.; Patel, K.; Pham, H.; Whitaker, M.; Anglin, O.; Kambhampati, A.K.; Milucky, J.; Chai, S.J.; Kirley, P.D.; Alden, N.B.; et al. Severity of Disease Among Adults Hospitalized with Laboratory-Confirmed COVID-19 Before and During the Period of SARS-CoV-2 B.1.617.2 (Delta) Predominance—COVID-NET, 14 States, January–August 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1513–1519.
- Dao, T.L.; Hoang, V.T.; Nguyen, N.N.; Delerce, J.; Chaudet, H.; Levasseur, A.; Lagier, J.C.; Raoult, D.; Colson, P.; Gautret, P. Clinical outcomes in COVID-19 patients infected with different SARS-CoV-2 variants in Marseille, France. Clin. Microbiol. Infect. 2021, 27, 1516.e1–1516.e6.




