
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ana Patrícia Cardoso | + 2422 word(s) | 2422 | 2022-01-17 10:11:18 | | | |
| 2 | Vivi Li | Meta information modification | 2422 | 2022-01-18 02:47:01 | | |
Video Upload Options
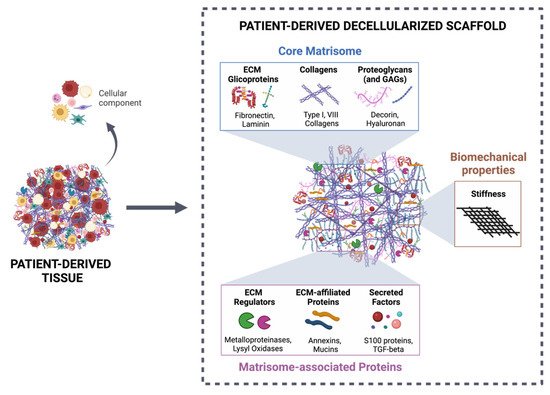
More than a physical structure providing support to tissues, the extracellular matrix (ECM) is a complex and dynamic network of macromolecules that modulates the behavior of both cancer cells and associated stromal cells of the tumor microenvironment (TME). Over the last few years, several efforts have been made to develop new models that accurately mimic the interconnections within the TME and specifically the biomechanical and biomolecular complexity of the tumor ECM. Particularly in colorectal cancer, the ECM is highly remodeled and disorganized and constitutes a key component that affects cancer hallmarks, such as cell differentiation, proliferation, angiogenesis, invasion and metastasis. Therefore, several scaffolds produced from natural and/or synthetic polymers and ceramics have been used in 3D biomimetic strategies for colorectal cancer research. Nevertheless, decellularized ECM from colorectal tumors is a unique model that offers the maintenance of native ECM architecture and molecular composition.
1. Introduction
2. Decellularized Colorectal Cancer Matrices as Bioactive Scaffolds for Modeling the Tumor Microenvironment
| ECM Sources | Decellularization Method | Biochemical Evaluation | Biomechanical Evaluation | REF |
|---|---|---|---|---|
| Cell-derived matrix HT-29 SW480 CCD-841-Com |
-CHEMICAL 0.5% Triton X-100 20 mM NH4OH Ionic and nonionic surfactants |
n/a | n/a | [32][33][34] |
| Human-derived tissue | CHEMICAL 5 mM EDTA 10% DMSO 1% Triton X-100 10 mM sodium cholate hydrate 50 mM Tris-HCl Centrifugal rotation Ionic and nonionic surfactants Mechanical mixing |
-Cellular proteins (cytokeratin, vimentin) and stromal components (collagen IV, fibrinogen, hyaluronic acid): Immunohistochemistry -Actin: Western Blot -DNA content: SYBR agarose gel |
-Architecture: HE -3D structure: FITC staining of ECMs |
[35] |
| CHEMICAL/ENZYMATIC 4% sodium deoxycholate 2000 kU DNase-I |
-DNA content: DNeasy Blood & Tissue kit -Stromal components (GAGs, Col IV): PAS and Immunohistochemistry -Cellular proteins (Ki67, vimentin, E-cadherin, DAPI): Immunofluorescence |
-Architecture: HE and Laminin -3D structure: SEM -Permeability: In-house developed permeability device |
[36] | |
| -DNA content: DNeasy Blood & Tissue kit and 1% SYBRsafe agarose gel -Stromal components (GAGs, Col IV): PAS, Masson’s Trichrome, Immunohistochemistry and Alcian blue |
-Architecture: HE, Gieson and Silver stains -3D structure: SEM |
[19] | ||
| PHYSICAL/CHEMICAL Freezing 2% SDC 1% Triton X-100 Physical disruption Ionic and nonionic surfactants |
-Nucleic acids: HE -Collagens: SHG |
-Stiffness: AMR -Topography: SHG |
[37] | |
| CHEMICAL/ENZYMATIC 0.1% SDS 50 U/mL DNase-I Ionic surfactant |
-Nucleic acids: DAPI -DNA content: PureLink Genomic DNA Mini Kit -Histomorphological analysis: HE and Masson’s Trichrome -Major ECM proteins (Collagens I and IV, Laminin, Fibronectin and Hyaluronic acid): Immunofluorescence |
-Stiffness: Rheology -3D structure: SEM |
[26] | |
| CHEMICAL 1% SDS 1% Triton X-100 |
-DNA content: Nanodrop -Major ECM proteins (GAGs, Collagen I, Laminin and fibronectin): Immunostaining -Cellular proteins: F-actin (cytoskeleton), DAPI and HE (nuclei acid) |
-Structure and architecture: SEM and TEM | [38] | |
| SISmuc (small intestine submucosa + mucosa from decellularized porcine jejunum) |
CHEMICAL 4% SDS 200 U/mL DNase I- |
n/e | n/e | [39] |
| Mice-derived tissue | CHEMICAL/ENZYMATIC 4% sodium deoxycholate 2000 kU DNase-I |
-DNA content: Roche’s DNA isolation Kit and Quant-It PicoGreen dsDNA Assay -Nucleic acids: DAPI and HE -Major ECM proteins (Collagens I and IV, Fibronectin and Laminin): Immunofluorescence and Masson’s Trichrome |
-Tensile testing: RSA-G2 solids analyzer | [40] |
References
- Dubé, C. Tackling colorectal cancer as a public health issue: What can the gastroenterologist do? Can. J. Gastroenterol. 2012, 26, 417–418.
- Vatandoust, S.; Price, T.J.; Karapetis, C.S. Colorectal cancer: Metastases to a single organ. World J. Gastroenterol. 2015, 21, 11767–11776.
- Choi, Y.; Sateia, H.F.; Peairs, K.S.; Stewart, R.W. Screening for colorectal cancer. Semin. Oncol. 2017, 44, 34–44.
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A., Jr. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375.
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell Sci. 2012, 125, 5591–5596.
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219.
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253.
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674.
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120.
- Scott, K.E.; Fraley, S.I.; Rangamani, P. A spatial model of YAP/TAZ signaling reveals how stiffness, dimensionality, and shape contribute to emergent outcomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2021571118.
- Sullivan, W.J.; Mullen, P.J.; Schmid, E.W.; Flores, A.; Momcilovic, M.; Sharpley, M.S.; Jelinek, D.; Whiteley, A.E.; Maxwell, M.B.; Wilde, B.R.; et al. Extracellular Matrix Remodeling Regulates Glucose Metabolism through TXNIP Destabilization. Cell 2018, 175, 117–132.e21.
- Park, J.S.; Burckhardt, C.J.; Lazcano, R.; Solis, L.M.; Isogai, T.; Li, L.; Chen, C.S.; Gao, B.; Minna, J.D.; Bachoo, R.; et al. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature 2020, 578, 621–626.
- Peyrou, M.; Clément, S.; Maier, C.; Bourgoin, L.; Branche, E.; Conzelmann, S.; Kaddai, V.; Foti, M.; Negro, F. PTEN protein phosphatase activity regulates hepatitis C virus secretion through modulation of cholesterol metabolism. J. Hepatol. 2013, 59, 420–426.
- You, Y.; Zheng, Q.; Dong, Y.; Wang, Y.; Zhang, L.; Xue, T.; Xie, X.; Hu, C.; Wang, Z.; Chen, R.; et al. Higher Matrix Stiffness Upregulates Osteopontin Expression in Hepatocellular Carcinoma Cells Mediated by Integrin β1/GSK3β/β-Catenin Signaling Pathway. PLoS ONE 2015, 10, e0134243.
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.T.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell 2009, 139, 891–906.
- Bays, J.L.; Campbell, H.K.; Heidema, C.; Sebbagh, M.; DeMali, K.A. Linking E-cadherin mechanotransduction to cell metabolism through force-mediated activation of AMPK. Nat. Cell Biol. 2017, 19, 724–731.
- Baker, B.M.; Chen, C.S. Deconstructing the third dimension—How 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012, 125, 3015–3024.
- Devarasetty, M.; Dominijanni, A.; Herberg, S.; Shelkey, E.; Skardal, A.; Soker, S. Simulating the human colorectal cancer microenvironment in 3D tumor-stroma co-cultures in vitro and in vivo. Sci. Rep. 2020, 10, 9832.
- Piccoli, M.; D’Angelo, E.; Crotti, S.; Sensi, F.; Urbani, L.; Maghin, E.; Burns, A.; De Coppi, P.; Fassan, M.; Rugge, M.; et al. Decellularized colorectal cancer matrix as bioactive microenvironment for in vitro 3D cancer research. J. Cell. Physiol. 2018, 233, 5937–5948.
- Kondo, J.; Ekawa, T.; Endo, H.; Yamazaki, K.; Tanaka, N.; Kukita, Y.; Okuyama, H.; Okami, J.; Imamura, F.; Ohue, M.; et al. High-throughput screening in colorectal cancer tissue-originated spheroids. Cancer Sci. 2019, 110, 345–355.
- Di Modugno, F.; Colosi, C.; Trono, P.; Antonacci, G.; Ruocco, G.; Nisticò, P. 3D models in the new era of immune oncology: Focus on T cells, CAF and ECM. J. Exp. Clin. Cancer Res. 2019, 38, 7.
- Sensi, F.; D’Angelo, E.; D’Aronco, S.; Molinaro, R.; Agostini, M. Preclinical three-dimensional colorectal cancer model: The next generation of in vitro drug efficacy evaluation. J. Cell. Physiol. 2018, 234, 181–191.
- Xu, R.; Zhou, X.; Wang, S.; Trinkle, C. Tumor organoid models in precision medicine and investigating cancer-stromal interactions. Pharmacol. Ther. 2021, 218, 107668.
- Hoshiba, T. Decellularized Extracellular Matrix for Cancer Research. Materials 2019, 12, 1311.
- Ferreira, L.P.; Gaspar, V.M.; Mano, J.F. Decellularized Extracellular Matrix for Bioengineering Physiomimetic 3D in Vitro Tumor Models. Trends Biotechnol. 2020, 38, 1397–1414.
- Pinto, M.L.; Rios, E.; Silva, A.C.; Neves, S.C.; Caires, H.R.; Pinto, A.T.; Durães, C.; Carvalho, F.A.; Cardoso, A.P.; Santos, N.C.; et al. Decellularized human colorectal cancer matrices polarize macrophages towards an anti-inflammatory phenotype promoting cancer cell invasion via CCL18. Biomaterials 2017, 124, 211–224.
- Wishart, A.L.; Conner, S.J.; Guarin, J.R.; Fatherree, J.P.; Peng, Y.; McGinn, R.A.; Crews, R.; Naber, S.P.; Hunter, M.; Greenberg, A.S.; et al. Decellularized extracellular matrix scaffolds identify full-length collagen VI as a driver of breast cancer cell invasion in obesity and metastasis. Sci. Adv. 2020, 6, eabc3175.
- Lv, Y.; Wang, H.; Li, G.; Zhao, B. Three-dimensional decellularized tumor extracellular matrices with different stiffness as bioengineered tumor scaffolds. Bioact. Mater. 2021, 6, 2767–2782.
- Liu, G.; Wang, B.; Li, S.; Jin, Q.; Dai, Y. Human breast cancer decellularized scaffolds promote epithelial-to-mesenchymal transitions and stemness of breast cancer cells in vitro. J. Cell. Physiol. 2019, 234, 9447–9456.
- Leiva, M.C.; Garre, E.; Gustafsson, A.; Svanström, A.; Bogestål, Y.; Håkansson, J.; Ståhlberg, A.; Landberg, G. Breast cancer patient-derived scaffolds as a tool to monitor chemotherapy responses in human tumor microenvironments. J. Cell. Physiol. 2021, 236, 4709–4724.
- Landberg, G.; Fitzpatrick, P.; Isakson, P.; Jonasson, E.; Karlsson, J.; Larsson, E.; Svanström, A.; Rafnsdottir, S.; Persson, E.; Gustafsson, A.; et al. Patient-Derived Scaffolds Uncover Breast Cancer Promoting Properties of the Microenvironment. Biomaterials 2020, 235, 119705.
- Hoshiba, T.; Tanaka, M. Decellularized matrices as in vitro models of extracellular matrix in tumor tissues at different malignant levels: Mechanism of 5-fluorouracil resistance in colorectal tumor cells. Biochim. Biophys. Acta (BBA)-Bioenerg. 2016, 1863, 2749–2757.
- Hoshiba, T.; Tanaka, M. Optimization of the tissue source, malignancy, and initial substrate of tumor cell-derived matrices to increase cancer cell chemoresistance against 5-fluorouracil. Biochem. Biophys. Res. Commun. 2015, 457, 353–357.
- Hoshiba, T. An extracellular matrix (ECM) model at high malignant colorectal tumor increases chondroitin sulfate chains to promote epithelial-mesenchymal transition and chemoresistance acquisition. Exp. Cell Res. 2018, 370, 571–578.
- Genovese, L.; Zawada, L.; Tosoni, A.L.; Ferri, A.; Zerbi, P.; Allevi, R.; Nebuloni, M.; Alfano, M. Cellular Localization, Invasion, and Turnover Are Differently Influenced by Healthy and Tumor-Derived Extracellular Matrix. Tissue Eng. Part A 2014, 20, 2005–2018.
- Sensi, F.; D’Angelo, E.; Piccoli, M.; Pavan, P.; Mastrotto, F.; Caliceti, P.; Biccari, A.; Corallo, D.; Urbani, L.; Fassan, M.; et al. Recellularized Colorectal Cancer Patient-Derived Scaffolds as In Vitro Pre-Clinical 3D Model for Drug Screening. Cancers 2020, 12, 681.
- Romero-López, M.; Trinh, A.; Sobrino, A.; Hatch, M.M.; Keating, M.T.; Fimbres, C.; Lewis, D.E.; Gershon, P.D.; Botvinick, E.L.; Digman, M.; et al. Recapitulating the human tumor microenvironment: Colon tumor-derived extracellular matrix promotes angiogenesis and tumor cell growth. Biomaterials 2017, 116, 118–129.
- Chen, H.J.; Wei, Z.; Sun, J.; Bhattacharya, A.; Savage, D.J.; Serda, R.; Mackeyev, Y.; Curley, S.A.; Bu, P.; Wang, L.; et al. A recellularized human colon model identifies cancer driver genes. Nat. Biotechnol. 2016, 34, 845–851.
- Nietzer, S.; Baur, F.; Sieber, S.; Hansmann, J.; Schwarz, T.; Stoffer, C.; Häfner, H.; Gasser, M.; Waaga-Gasser, A.M.; Walles, H.; et al. Mimicking Metastases Including Tumor Stroma: A New Technique to Generate a Three-Dimensional Colorectal Cancer Model Based on a Biological Decellularized Intestinal Scaffold. Tissue Eng. Part C Methods 2016, 22, 621–635.
- Alabi, B.R.; LaRanger, R.; Shay, J.W. Decellularized mice colons as models to study the contribution of the extracellular matrix to cell behavior and colon cancer progression. Acta Biomater. 2019, 100, 213–222.
- Taylor, D.A.; Sampaio, L.C.; Ferdous, Z.; Gobin, A.S.; Taite, L.J. Decellularized matrices in regenerative medicine. Acta Biomater. 2018, 74, 74–89.
- Park, Y.; Huh, K.M.; Kang, S.-W. Applications of Biomaterials in 3D Cell Culture and Contributions of 3D Cell Culture to Drug Development and Basic Biomedical Research. Int. J. Mol. Sci. 2021, 22, 2491.
- Dzobo, K.; Motaung, K.; Adesida, A. Recent Trends in Decellularized Extracellular Matrix Bioinks for 3D Printing: An Updated Review. Int. J. Mol. Sci. 2019, 20, 4628.
- Zhang, W.; Du, A.; Liu, S.; Lv, M.; Chen, S. Research progress in decellularized extracellular matrix-derived hydrogels. Regen. Ther. 2021, 18, 88–96.
- Schanz, J.; Pusch, J.; Hansmann, J.; Walles, H. Vascularised human tissue models: A new approach for the refinement of biomedical research. J. Biotechnol. 2010, 148, 56–63.
- Tian, X.; Werner, M.E.; Roche, K.C.; Hanson, A.D.; Foote, H.P.; Yu, S.K.; Warner, S.B.; Copp, J.A.; Lara, H.; Wauthier, E.L.; et al. Organ-specific metastases obtained by culturing colorectal cancer cells on tissue-specific decellularized scaffolds. Nat. Biomed. Eng. 2018, 2, 443–452.
- Naba, A.; Clauser, K.R.; Ding, H.; Whittaker, C.A.; Carr, S.A.; Hynes, R.O. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 2016, 49, 10–24.
- D’Angelo, E.; Natarajan, D.; Sensi, F.; Ajayi, O.; Fassan, M.; Mammano, E.; Pilati, P.; Pavan, P.; Bresolin, S.; Preziosi, M.; et al. Patient-Derived Scaffolds of Colorectal Cancer Metastases as an Organotypic 3D Model of the Liver Metastatic Microenvironment. Cancers 2020, 12, 364.
- Parkinson, G.T.; Salerno, S.; Ranji, P.; Håkansson, J.; Bogestål, Y.; Wettergren, Y.; Ståhlberg, A.; Bexe Lindskog, E.B.; Landberg, G. Patient-derived scaffolds as a model of colorectal cancer. Cancer Med. 2021, 10, 867–882.
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017, 8, 077.
- Sanz-Pamplona, R.; Berenguer, A.; Cordero, D.; Molleví, D.G.; Crous-Bou, M.; Sole, X.; Paré-Brunet, L.; Guino, E.; Salazar, R.; Santos, C.; et al. Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer. Mol. Cancer 2014, 13, 46.
- Ahmed, E.; Saleh, T.; Xu, M. Recellularization of Native Tissue Derived Acellular Scaffolds with Mesenchymal Stem Cells. Cells 2021, 10, 1787.
- Gustafsson, A.; Garre, E.; Leiva, M.C.; Salerno, S.; Ståhlberg, A.; Landberg, G. Patient-derived scaffolds as a drug-testing platform for endocrine therapies in breast cancer. Sci. Rep. 2021, 11, 3334.




