
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mohmed Isaqali Karobari | + 1374 word(s) | 1374 | 2021-12-29 11:51:43 | | | |
| 2 | Yvaine Wei | Meta information modification | 1374 | 2021-12-30 03:26:41 | | |
Video Upload Options
Antimicrobial resistance (AMR) is a global health issue that plays a significant role in morbidity and mortality, especially in immunocompromised patients. It also becomes a serious threat to the successful treatment of many bacterial infections. The widespread and irrelevant use of antibiotics in hospitals and local clinics is the leading cause of AMR.
1. Introduction
COVID-19 infections have far exceeded bacterial co-infection and mortality rates compared to other common respiratory viral infections [1]. The co-infection of SARS-CoV-2 with other microbes, mainly bacteria and fungus, is a determining factor in COVID-19 development, making diagnosis, treatment, and prognosis more complicated. In individuals with COVID-19, bacterial co-infection has been linked to disease progression and prognosis. This scenario increases the need for critical care units, antibiotic therapy, and mortality [2]. Unfortunately, due to their widespread use, human may face the emergence of multi-drug resistant (MDR) pathogens leading to reduced efficacy of most potent antimicrobials [2][3]. AMR is a global problem that poses a severe threat to the success of treating a wide range of bacterial infections and affects many hospitalised patients, and most probably becomes a serious threat to the patients who are admitted to the SICUs [4][5].
2. Antibiotic Susceptibility Patterns of Clinical Isolates

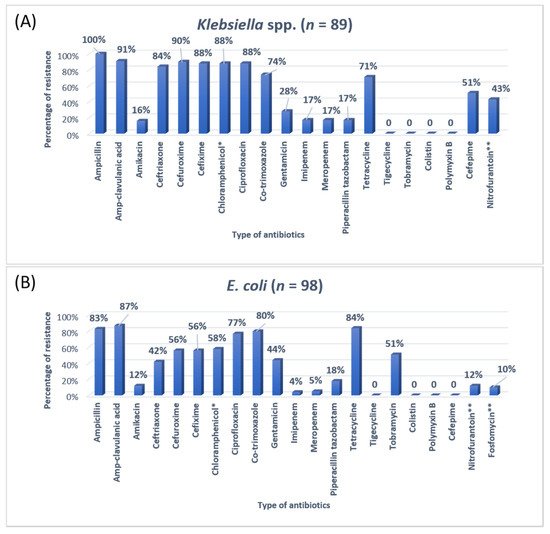
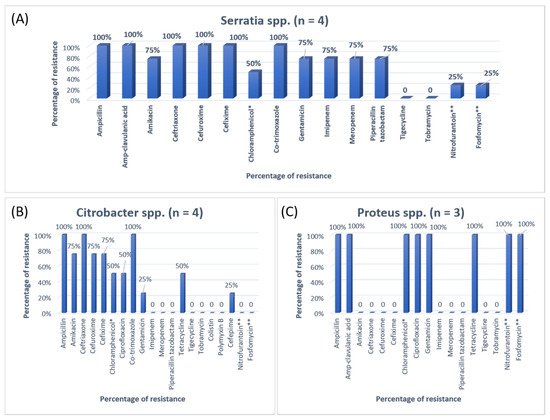
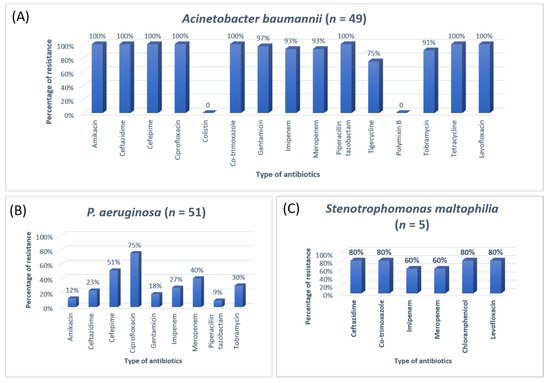
3. Research Findings
Antibiotics are the most commonly prescribed drugs among hospitalised patients, especially in SICUs. It is imperative to use the appropriate antibiotics in intensive care units with few prescriptions as an acceptable quality of care, infection control, cost reduction, and length of hospital stay [15][16][17]. Patients admitted in the SICU are critically ill requiring prescribed the medicine without waiting for the culture reports that give information about the antimicrobial resistance pattern of the suspected organism for a specific cause [18][19]. In terms of the culture reports, there is no possibility to wait for reports because these take a minimum of 48 h, and as a result antibiotic resistance occurs in patients admitted to the SICUs [20]. The current study was conducted among COVID-19 patients admitted in SICUs of tertiary care hospitals, requiring monitoring and special care to analyse the antibiotics utilisation pattern and determine the prevalence of AMR.
A previous study on microbial infection and antibiotic resistance patterns in COVID-19 patients admitted in SICUs of tertiary care hospitals showed that Pseudomonas was the most common organism identified in the medical ICU, followed by Klebsiella pneumonia [21]. A study on the prevalence of microorganisms and bacterial resistance in the SICU of the Bangabandhu Sheikh Mujib Medical University of Bangladesh showed that the maximum identified organism was Acetobacter. (45.4%), of P. aeruginosa (32.2%), Proteus (11%), Klebsiella pneumoniae 10%, and E. coli (3%) were identified [22]. A study by Mehta et al., 2015 on ICU patients revealed that the Pseudomonas spp. (29.1%) was the most common organism, followed by Acinetobacter spp. (27.5%) [16]. Another previous analysis of AST and bacteriology profile on patients at tertiary care hospitals in Ahmadabad showed that Acinetobacter spp. [30.9%] was the most common organism, after coming Klebsiella spp. (29.8%) and P. aeruginosa (22.9%) [17]. However, in the current study, the most common isolated organism was E. coli (38%), followed by Klebsiella pneumoniae (24%), P. aeruginosa (14%). While Streptococcus agalactiae, Citrobacter freundii, Serratia liqeuficiens, and Stenotrophomonas maltophilla were 1.7%, 1.1%, 1.1% and 1.4%, respectively.
4. Conclusions
References
- Mondal, M.K.; Roy, B.R.; Yeasmeen, S.; Haque, F.; Huda, A.Q.; Banik, D. Prevalence of microorganism and emergence of bacterial resistance in ICU of Bangabandhu Sheikh Mujib Medical University of Bangladesh. J. Bangladesh Soc. Anaesthesiol. 2013, 26, 20–26.
- Parveen, S.; Saqib, S.; Ahmed, A.; Shahzad, A.; Ahmed, N. Prevalence of MRSA colonisation among healthcare-workers and effectiveness of decolonisation regimen in ICU of a Tertiary care Hospital, Lahore, Pakistan. Adv. Life Sci. 2020, 8, 38–41.
- Ventola, C.L. The antibiotic resistance crisis: Part 2: Management strategies and new agents. Pharm. Ther. 2015, 40, 344.
- Pickens, C.I.; Wunderink, R.G. Principles and practice of antibiotic stewardship in the ICU. Chest 2019, 156, 163–171.
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance—The need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098.
- Smith, R.; Coast, J. The true cost of antimicrobial resistance. BMJ 2013, 346, 1–5.
- Dyar, O.J.; Castro-Sánchez, E.; Holmes, A.H. What makes people talk about antibiotics on social media? A retrospective analysis of Twitter use. J. Antimicrob. Chemother. 2014, 69, 2568–2572.
- Knight, G.M.; Glover, R.E.; McQuaid, C.F.; Olaru, I.D.; Gallandat, K.; Leclerc, Q.J.; Fuller, N.M.; Willcocks, S.J.; Hasan, R.; van Kleef, E.; et al. Antimicrobial resistance and COVID-19: Intersections and implications. Elife 2021, 10, e64139.
- Monnet, D.L.; Harbarth, S. Will coronavirus disease (COVID-19) have an impact on antimicrobial resistance? Eurosurveillance 2020, 25, 2001886.
- Ali, Z.; Jatoi, M.A.; Al-Wraikat, M.; Ahmed, N.; Li, J. Time to Enhance Immunity via Functional Foods and Supplements: Hope for SARS-CoV-2 Outbreak. Altern. Ther. Health Med. 2020, 27, 30–44.
- Adiga, M.S.; Alwar, M.; Pai, M.; Adiga, U.S. Pattern of antimicrobial agents use in hospital deliveries: A prospective comparative study. Online J. Health Allied Sci. 2010, 8, 10.
- Vincent, J.-L.; Rello, J.; Marshall, J.; Silva, E.; Anzueto, A.; Martin, C.D.; Moreno, R.; Lipman, J.; Gomersall, C.; Sakr, Y.; et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA 2009, 302, 2323–2329.
- Zahra, N.; Zeshan, B.; Qadri, M.M.A.; Ishaq, M.; Afzal, M.; Ahmed, N. Phenotypic and Genotypic Evaluation of Antibiotic Resistance of Acinetobacter baumannii Bacteria Isolated from Surgical Intensive Care Unit Patients in Pakistan. Jundishapur J. Microbiol. 2021, 14, e113008.
- Bataineh, H.A.; Alrashed, K.M. Resistant gram-negative bacilli and antibiotic consumption in Zarqa, Jordan. Pak. J. Med. Sci. 2007, 23, 59–63.
- Ahmed, N.; Ali, Z.; Riaz, M.; Zeshan, B.; Wattoo, J.I.; Aslam, M.N. Evaluation of Antibiotic Resistance and Virulence Genes among Clinical Isolates of Pseudomonas aeruginosa from Cancer Patients. Asian Pac. J. Cancer Prev. APJCP 2020, 21, 1333–1338.
- Mehta, T.; Chauhan, B.; Rathod, S.; Pethani, J.; Shah, P.D. Bacterilogical profile and drug resistance pattern of isolates of the patients admitted in medical intensive care unit of a tertiary care hospital in Ahmedabad. Med. Sci. 2015, 4, 222–225.
- Barai, L.; Fatema, K.; Haq, J.A.; Faruq, M.O.; Ahsan, A.A.; Morshed, M.A.H.G.; Hossain, M.B. Bacterial profile and their antimicrobial resistance pattern in an intensive care unit of a tertiary care hospital of Dhaka. Ibrahim Med. Coll. J. 2010, 4, 66–69.
- Lockhart, S.R.; Abramson, M.A.; Beekmann, S.E.; Gallagher, G.; Riedel, S.; Diekema, D.J.; Quinn, J.P.; Doern, G.V. Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J. Clin. Microbiol. 2007, 45, 3352–3359.
- Liang, S.Y.; Kumar, A. Empiric antimicrobial therapy in severe sepsis and septic shock: Optimising pathogen clearance. Curr. Infect. Dis. Rep. 2015, 17, 36.
- Ahmed, N.; Zeshan, B.; Naveed, M.; Afzal, M.; Mohamed, M. Antibiotic resistance profile in relation to virulence genes fimH, hlyA and usp of uropathogenic E. coli isolates in Lahore, Pakistan. Trop. Biomed. 2019, 36, 559–568.
- Sanjana, R.; Shah, R.; Chaudhary, N.; Singh, Y. Prevalence and antimicrobial susceptibility pattern of methicillin-resistant Staphylococcus aureus (MRSA) in CMS-teaching hospital: A preliminary report. J. Coll. Med. Sci.-Nepal 2010, 6, 1–6.
- Radji, M.; Fauziah, S.; Aribinuko, N. Antibiotic sensitivity pattern of bacterial pathogens in the intensive care unit of Fatmawati Hospital, Indonesia. Asian Pac. J. Trop. Biomed. 2011, 1, 39–42.
- Tuem, K.B.; Desta, R.; Bitew, H.; Ibrahim, S.; Hishe, H.Z. Antimicrobial resistance patterns of uropathogens isolated between 2012 and 2017 from a tertiary hospital in Northern Ethiopia. J. Glob. Antimicrob. Resist. 2019, 18, 109–114.
- Mohamad, N.A.; Jusoh, N.A.; Htike, Z.Z.; Win, S.L. Bacteria identification from microscopic morphology: A survey. Int. J. Soft Comput. Artif. Intell. Appl. (IJSCAI) 2014, 3, 1–12.
- Phillips-Jones, M.K.; Harding, S.E. Antimicrobial resistance (AMR) nanomachines—Mechanisms for fluoroquinolone and glycopeptide recognition, efflux and/or deactivation. Biophys. Rev. 2018, 10, 347–362.
- Yang, J.; Zheng, Y.; Gou, X.; Pu, K.; Chen, Z.; Guo, Q.; Ji, R.; Wang, H.; Wang, Y.; Zhou, Y. Prevalence of comorbidities in the novel Wuhan coronavirus (COVID-19) infection: A systematic review and meta-analysis. Int. J. Infect. Dis. 2020, 10, 91–95.




